-
PDF
- Split View
-
Views
-
Cite
Cite
Shazia Firdous, Abhirupa Ghosh, Sudipto Saha, BCSCdb: a database of biomarkers of cancer stem cells, Database, Volume 2022, 2022, baac082, https://doi.org/10.1093/database/baac082
Close - Share Icon Share
Abstract
Cancer stem cells (CSCs) are a small heterogeneous population present within the tumor cells exhibiting self-renewal properties. CSCs have been demonstrated to elicit an important role in cancer recurrence, metastasis and drug resistance. CSCs are distinguished from cancer cell populations based on their molecular profiling or expression of distinct CSC biomarker(s). Recently, a huge amount of omics data have been generated for the characterization of CSCs, which enables distinguishing CSCs in different cancers. Here, we report biomarkers of the Cancer Stem Cells database (BCSCdb), a repository of information about CSC biomarkers. BCSCdb comprises CSC biomarkers collected from PubMed literature where these are identified using high-throughput and low-throughput methods. Each biomarker is provided with two different scores: the first is a confidence score to give confidence to reported CSC biomarkers based on the experimental method of detection in CSCs. The second is the global score to identify the global CSC biomarkers across 10 different types of cancer. This database contains three tables containing information about experimentally validated CSC biomarkers or genes, therapeutic target genes of CSCs and CSC biomarkers interactions. It contains information on three types of markers: high-throughput marker (HTM-8307), high-throughput marker validated by the low-throughput method (283) and low-throughput marker (LTM-525). A total of 171 low-throughput biomarkers were identified in primary tissue referred to as clinical biomarkers. Moreover, it contains 445 target genes for CSC therapeutics, 10 biomarkers targeted by clinical trial drugs in CSCs and 5 different types of interaction data for CSC biomarkers. BCSCdb is an online resource for CSC biomarkers, which will be immensely helpful in the cancer research community and is freely available.
Database URL: http://dibresources.jcbose.ac.in/ssaha4/bcscdb
Introduction
Cancer stem cells (CSCs) contribute to cancer recurrence, metastasis, evasion of immunological surveillance and resistance to chemotherapy and radiotherapy (1–5). Due to these mentioned features, CSCs are considered one of the main targets of novel experimental and cancer therapeutics (6, 7). Identifying and isolating CSCs is the preliminary stage in CSC-based therapeutic design and improving the therapeutic efficacy of cancer treatment. The stemness of CSCs is supported by both intracellular and extracellular signals, including the molecular pathways, surrounding niche and transcriptional factors (8). CSCs were first isolated with the help of surface marker expression, thereby to date many surface and intracellular CSC biomarkers have been evaluated to purify CSCs from heterogeneous cancer cells (9). Many different surface markers such as CD133, CD44 and EpCAM have been identified as potential CSC markers in many solid and nonsolid tumors. These transmembrane surface biomarkers have been recognized as a prominent therapeutic target to eliminate the CSC’s therapeutic resistance in different types of cancer (10–12). Besides, activation of many intracellular pluripotency factors such as SOX2, NANOG and POU5F1 has also been reported to regulate the stemness properties and serve as a vital CSC biomarker (13, 14). Moreover, researchers are now exploring the molecular profiling of CSCs by applying the transcriptomic and proteomic approaches and have identified functional CSC biomarker-related genes and regulating pathways (15, 16). Furthermore, uncovering regulatory interactions specific to CSCs is crucial for illustrating the complex molecular mechanism inside the CSC population that led to gaining the stemness properties. Also, the deep insight of the CSC interaction network opens the door to designing network-based therapeutic approaches that enable targeting the entire regulatory network instead of targeting a single element (17).
As the CSC contribution to cancer resistance has been supported by an ample amount of data, investigations have been performed on CSCs. A few databases have been published till now dedicated to CSCs to enhance the further knowledge of cancer therapeutics. CSCdb: a portal of cancer stem cell for markers, related genes and functional information provides literature-based information about CSC marker, CSC-related genes and their functional annotation from tissues. Another database, CSCTT: CSCs therapeutic target database, provides validated therapeutic targets for CSCs, but the URL of both the databases are not working now and only the literature content is available online (18, 19). To date, omics approaches were applied for identifying novel CSC biomarkers for different types of cancer, and these studies resulted in the generation of a huge number of markers regulating CSC properties. This CSC-related biomarker information is scattered in plenty of literature. Therefore, there is an urgent need for a bioinformatics resource that acts as a useful tool for CSC researchers.
BCSCdb is a manually curated database of high-throughput markers (HTMs) and low-throughput markers (LTMs) for CSCs, collected from PubMed literatures. At present, BCSCdb contains CSC biomarkers from cancer cell lines and primary tissues that are obtained from 10 different types of CSCs, inclusive of their therapeutic target gene and interaction data if available. Each biomarker is provided with two different scores: the first is the confidence score and the second is the global score. The confidence scoring system was performed for the experimentally validated method, and it helps in assessing the confidence of a CSC biomarker in the CSC population. The global scoring system was performed based on the frequency of a CSC biomarker across 10 different types of cancer reported in BCSCdb; this will aid in identifying global CSC biomarkers and local CSC biomarkers specified to a particular type of cancer.
Materials and methods
Data procurement of BCSCdb
The information about the CSC biomarker was manually collected (till May 2022) and curated from the published scientific literature. To find out the CSC-related biomarker articles for different types of cancer, a query in PubMed advanced search is made using multiple keywords such as (Lung/Breast/Colon/Glioma/Head and Neck/Melanoma/Bladder/Hepatic/Pancreatic/Gastric) [Title] AND cancer [Title] AND stem [Title/Abstract]. Similarly, to find out a particular information about the molecular interaction of CSCs, a query in PubMed advanced search is made using a combination keyword such as Interaction[Title] AND cancer [Title] AND stem [Title/Abstract]. The animal cell line-specific papers, review papers and non-English papers were removed and the information about the CSC biomarkers from human primary tissues and human cell lines was collected for different types of cancer. A total of 1962 scientific works of literature were curated, and extensive literature mining was performed to obtain experimentally validated information about CSC biomarkers and their therapeutic approaches, along with the molecular interaction data if available.
Data architecture of BCSCdb and implementation
The CSC biomarker, CSC therapeutics and CSC interaction data were stored in the form of the table and were converted to structured data. The BCSCdb is implemented using the Apache HTTP 2.2.15 web server and the MySQL 5.1.69 database server. The web interface has been designed with PHP 5.3.3, HTML, JavaScript and CSS. In BCSCdb, the high-throughput and low-throughput biomarkers of CSCs, therapeutic targets of CSCs, and interactor genes or protein or microRNA (miRNA) were hyperlinked with their respective HGNC IDs (https://www.genenames.org/) (20). The drugs or the small-molecule inhibitors were linked with PubChem IDs (https://pubchem.ncbi.nlm.nih.gov/) (21). In addition, the reference ID provides a link to PubMed. Supplementary file 1, Figure S1 depicts the BCSCdb implementation and a summary statistic of the database.
Biomarker scoring method
The experimentally validated cancer cell line-specific and primary tissue-specific biomarkers that are used to identify the CSC populations were compiled based on their detection method and functional method. In order to make the user get comprehensive information about CSC biomarkers, we further classified biomarkers as HTM and LTM. The HTM were collected from transcriptomic and proteomic studies, and LTM markers were collected from detection and isolation methods, such as reverse transcriptase-polymerase chain reaction (RT-PCR), western blotting, immunohistochemistry (IHC) staining and fluorescence-activated cell sorting (FACS). Two kinds of scoring systems have been developed for each CSC biomarker by taking two parameters into consideration such as the biomarker identification method in a particular type of cancer or cell line named as the confidence score and the frequency of a CSC biomarker in BCSCdb in 10 different types of cancer named as the global score. The formulae that have been used to compute the scores for the CSC biomarkers are listed in the sections “Confidence score” and “Global score”.
Confidence score
The weightage of the confidence score has been selected based on their identification method as given in Supplementary file 1, Table ST1.
For cell line-based studies, western blotting has been given the highest score of 0.7, and transcriptomics has been given the lowest score of 0.1 (22, 23). A 0.2 score has been added to each method for primary tissue, where 0.9 is the highest score in the case of primary tissue applied for western blotting and 0.3 is the lowest score used for transcriptomics.
For example, if a biomarker is detected using western blotting techniques, then it is more likely to confirm its expression at the protein level. Similarly, if a biomarker is detected using transcriptomic study, it is assigned the lowest-scoring value since it is at the mRNA level. The confidence scores were assigned using the following formula:
where |${\rm{C}}{{\rm{S}}_{\rm{A}}}$| is the confidence score of gene A and |${\rm{M}}{{\rm{S}}_{\rm{i}}}$| is the method score as assigned in Supplementary file 1, Table ST1.
The scoring values were normalized to a scale of 0.04 to 1 by using the following formula:
where |${\rm{Maximum}}_{{\rm{i}} = 1}^{\rm{n}}\left( {{\rm{C}}{{\rm{S}}_{\rm{A}}}} \right)$| is 2.5, the highest combinatorial method score observed in BCSCdb. As the detection method for a CSC biomarker specific to a cell line varies in publications, the highest method score was considered as the final confidence score for that biomarker in that cell line. For example, in the X article if ‘PROM1’ detection has been done by using the western blotting technique in the A549 cell line and in the Y article if ‘PROM1’ detection has been done by using the RT-PCR technique in the A549 cell line, then we have considered the western blotting score for every ‘PROM1’ in the A549 cell line entries in the database. Similarly, for primary tissue entries, we considered the highest method score allotted to a biomarker in a particular type of cancer as its final confidence score. The normalized confidence score for each unique detection method is given in Supplementary file 2, Table ST5. A normal distribution plot and the density plot of the confidence scores are provided in Supplementary file 1, Figure S2A and B. Furthermore, to obtain the confident CSC biomarkers, three threshold values were selected from the normal distribution plot: biomarkers having a confidence score of |$ \ge $|0.6 were recognized as high-confident CSC biomarkers. Biomarkers having a confidence score of 0.6–0.4 are considered moderate-confident biomarkers, and all the biomarkers having scores of 0.4–0.2 are recognized as the lowest confident biomarkers.
Global score
Global scoring denotes the frequency of a gene or a biomarker in the whole database.
where GSA is the global scoring of gene A, NA is the number of unique papers reporting gene A and NT is the number of unique papers.
The global scores were normalized to the scale of 0.001 to 1 using the following formula:
where |${\rm{G}}{{\rm{S}}_{\rm{i}}}$| is the global score of i-th gene.
The normalized global scores for each biomarker are provided in Supplementary file 2, Table ST6. A normal distribution plot and a density plot were made as shown in Supplementary file 1, Figure S3A and B. To obtain the relevant global CSC biomarkers, three thresholds, i.e. high (0.1), moderate (0.01) and low (<0.01) values, were selected.
Database content
The database structured three tables for CSC information: (i) CSC biomarker table, (ii) CSC biomarker interaction table and (iii) CSC therapeutic table. The biomarker table provides information on CSC biomarker gene names, marker type and the expression level of HTM markers along with the scores of biomarkers. The therapeutic table contains information about the therapeutic strategies used to target the CSC population and target gene names. It also mentioned the effects of the therapeutic approach on the CSC population, such as self-renewal, migration and apoptosis. The interaction table contains experimentally validated low-throughput cancer cell line-specific molecular interaction data obtained from CSCs. It includes five different types of interaction data from different types of CSC populations, including protein–protein interaction, protein–gene interaction, gene–gene interaction and miRNA–mRNA interaction. To construct a regulatory network for CSC molecular interaction, Cytoscape version 3.8.2 was used (24). The attributes of all tables are provided in Supplementary file 1, Figure S4.
Results
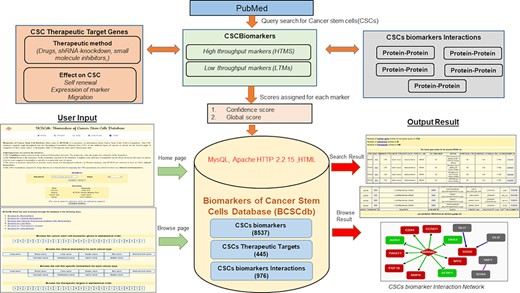
BCSCdb is designed as a user-friendly database, and it serves as a useful tool to collect information about HTMs and LTMs of CSCs and their respective therapeutic approaches and interaction networks. A schematic representation of BCSCdb with basic data statistics is shown in Figure 1.
Schematic representation of the workflow of BCSCdb online database and summary of data statistics.
Data statistics of biomarkers in BCSCdb
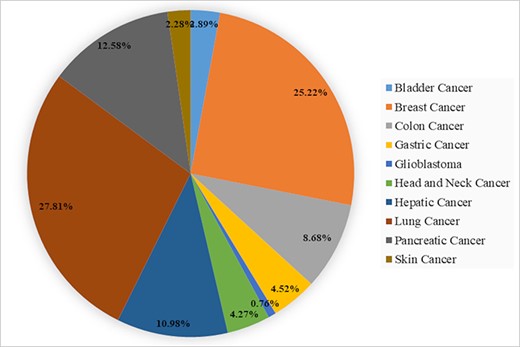
The data statistics of biomarkers in BCSCdb are represented in the form of a pie chart as shown in Figure 2 and Supplementary file 1, Tables ST2–ST4. The BCSCdb provides a significant amount of data for lung, breast and pancreatic cancer, and among lung cancer, the maximum number of entries are present for A549 and H2170 cell lines. For breast and pancreatic cancer, a notable number of data are present for MCF7, MDA-MB-231 and PANC-1 cell lines. A total of 171 clinical biomarkers were identified in primary tissues, and 10 clinical trial biomarkers were also reported. Besides this, it also provides 63 chemotherapeutic drug names that are responsible for enriching the CSC population. The therapeutic data contain 445 target genes and 383 drugs inhibiting CSC properties, such as self-renewal and migration invasion. It also provides information on CSC clinical trial drugs; here 11 clinical trial drugs were reported for all types of cancer reported in the BCSCdb.
A pie chart showing the percentage of CSCs biomarkers in 10 different types of cancer present in BCSCdb.
Search, browse and download options
On the ‘home page’, a brief introduction to the database is given. The home page has search and advanced search options for user queries based on keywords. The database can be queried by using the following keywords: Cell line, Method, Cancer type, Marker type and Marker name or can retrieve the data using ‘ALL’ (default option). Users can customize their search by using advanced search where logical operators (AND/OR/NOT) are implemented to get more comprehensive information for CSC biomarkers. The advanced search also helps to speed up some aspects of searching for CSC information. The search and advance search options are shown in Supplementary file 1, Figure S5A and B.
The browse page of BCSCdb allows users to browse the data in tabular format, and six different browse options are available at the top of the browse page. Snapshots of all the browse options are provided in Supplementary file 1, Figure S6A and B. First, it allows users to browse CSC biomarker targets in the alphabetical order of gene names. Second, the list of clinical biomarkers can be browsed by cancer type. The CSC biomarkers that are identified in primary tissue using low-throughput methods such as western blotting, RT-PCR and IHC staining are regarded as clinical biomarkers. Third, a browse option is available for drugs in clinical trials against CSC biomarker(s). A snapshot of this table is provided in Supplementary file 1, Figure S7. Fourth, the browse page also allows users to browse cancer cell line-specific CSC biomarkers. Fifth, users can also browse CSCs’ therapeutic targets in alphabetical order. Finally, the cell line-specific interaction networks can be retrieved in tabular format and can be visualized in regulatory network format. Furthermore, all the datasets of BCSCdb can be downloaded freely, including the CSC biomarkers and respective therapeutic information and interaction data. Users can explore the download option and save the datasets in CSV format for future reference.
Information on the output page
The output page for each record shows data from all three tables including biomarker, therapeutic and interaction. The biomarker dataset contains information about biomarker gene names, type of biomarker as HTM or LTM and the differential expression level of HTM, such as upregulated or downregulated in CSCs. In addition, it also provides information about cancer type, histological type, cell line, drugs used to enrich the CSC population, the functional and detection method for CSC isolation and reference ID. Furthermore, each CSC biomarker has its corresponding confidence score and global score. The biomarkers’ names are ranked based on the descending order of the confidence score.
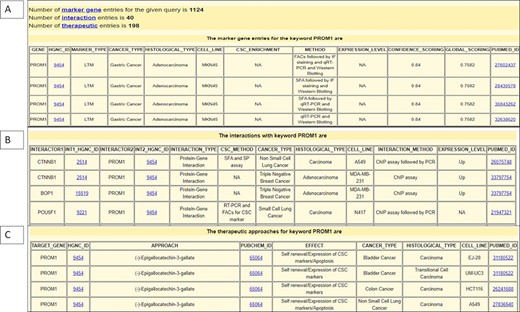
Similarly, the therapeutic dataset encompasses the information about drugs used to target CSC biomarkers with their respective PubChem IDs. It also provides information about therapeutic approaches including the biotherapy method, the therapeutic target gene name and the effect of the therapeutic approach on the CSC population, cancer type, histological type, cell line and reference ID. The interaction dataset compiles information about interactor genes with their respective HGNC ID, cancer type, histological type, cell line, CSC isolation method, interaction type, interaction method, expression level and reference ID. The total number of records generated for each input query is provided at the top of the output page. Figure 3 shows parts of the output page generated by querying the BCSCdb by putting ‘PROM1’ in the keyword search option and the key set as ‘marker’.
Snapshot of output page of BCSCdb showing the result of three tables (A) CSC biomarker, (B) CSC biomarker interaction and (C) CSC therapeutics, respectively, of query search ‘PROM1’.
Confident CSC biomarkers
The confidence score of CSC biomarkers helps to identify the confident biomarker by their experimental identification method as each identification method is provided with a weightage based on its efficiency (Supplementary file 1, Table ST1). For a confidence score, a threshold value of ≥0.6 is a high confidence score that gives a hit of 49 genes; by selecting a threshold value of 0.6–0.4, which is a medium confidence score, it gives a hit of 200 genes; and upon selecting a threshold value of ≥0.2 for a confidence score, which is the lowest confidence threshold value, it gives a hit of 838 genes. The confident biomarker list is provided in Supplementary file 2, Table ST7. Furthermore, we have identified 287 biomarkers, which include several miRNAs such as MIR151A, MIR320D1 and MIR221, and several unique genes ANXA1, BMP7 and FGF9 and interleukins IL6 and IL8—the expression of which was validated by high-throughput method followed by low-throughput method in different types of cancer. As these markers’ confidence scores are high, they can be further explored as a CSC therapeutic target. The list of markers that have been identified by using only high-throughput studies and only low-throughput methods, and markers identified by using both methods are provided in Supplementary file 2, Table ST8. The global score indicates the probability of a CSC biomarker to be considered a unique or global marker for 10 different types of cancer. Upon selecting a threshold value of 0.1 for global scoring, it gives a hit of 11 genes. Similarly, the medium confidence score of 0.01 gives a hit of 56 genes. The gene list that is obtained after considering the threshold value is provided in Supplementary file 2, Table (ST9). The top four surface markers that we get after giving a threshold value of 0.1 are CD44, EpCAM, PROM1 and ABCG2, and the intracellular markers include POU5F1, NANOG, SOX2 and BMI1. Furthermore, we have checked the presence of high and moderated scored global CSC biomarkers across the 10 types of cancer. The result is provided in Supplementary file 2, Table S10.
Case study for DCLK1 biomarkers using BCSCdb in colon and pancreatic cancer
We analyzed the data in BCSCdb for DCLK1 by performing a simple keyword search for ‘DCLK1’ with all options on the home page of BCSCdb. The result shows that the confidence score of DCLK1 is high in colon cancer (0.6), moderate in pancreatic (0.4) and low in lung cancer (0.04) but the global score of DCLK1 is very low (0.0159), which means that this biomarker is very specific to colon and pancreatic cancer. In BCSCdb, it is reported that inhibition of DCLK1 by using various therapeutic approaches in colon cancer as well as in pancreatic cancer can lead to a significant decrease in the self-renewal properties of CSCs. In addition, DCLK1 knockdown is responsible for reducing the expression of other stemness markers, including SOX2, NANOG and CD44. This case study thus shows that BCSCdb provides information for detecting potential therapeutic targets for CSCs and could be used to study the specificity of CSC biomarkers for a specific type of cancer.
Case study of MIR582 using BCSCdb interaction network in lung cancer
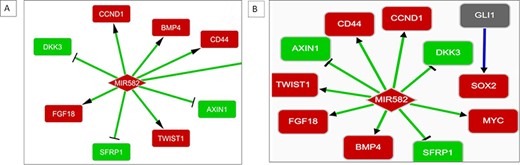
Upregulated expression of MIR582 has been observed in CSCs from a non-small-cell lung cancer (NSCLC) cell line A549 with a confidence score of 0.2 in BCSCdb. The interaction regulatory networks of two NSCLC cell lines, A549 and H1975, in BCSCdb have shown that the upregulated expression of MIR582 enhances the expression of crucial CSC-related genes CD44, CCND1 and MYC. It was also observed that it targets the WNT pathway antagonists, such as DKK3, SFRP1 and AXIN. The activation of the WNT pathway mediated by MIR582 is responsible for maintaining the stemness of cancer cells. Therefore, designing a therapeutic approach that can target MIR582 can directly disrupt the entire regulatory network maintained by WNT pathway genes, as shown in Figure 4A and B.
(A) Interaction network showing MIR582 regulation in the A549 NSCLC cell line. (B) Interaction network showing MIR582 regulation in the H1975 NSCLC cell line.
Comparison with the existing CSC databases
At present, as far as we know, two CSC-related databases are available. The CSCdb, a cancer stem cell portal for the marker, related genes and functional information, is a literature-based database mainly focused on CSC markers and functionally related CSC genes annotation (18). Another, the CSCTT, cancer stem cell therapeutic target database, deals with the CSC therapeutic target genes and drugs targeting CSCs (19). The content of these two databases was thoroughly studied and compared with the BCSCdb, as shown in Table 1. The unique attributes that are present in BCSCdb in comparison with CSCdb and CSCTT include cell line, histological type, CSC detection method, global score, confidence score, CSC enrichment drug and the effect of the drug on CSC trait.
Comparison of attributes and data content of BCSCdb with CSCdb and CSCTT
| Attributes . | BCSCdb . | CSCdb . | CSCTT . |
|---|---|---|---|
| Validation of biomarkers in the clinical sample | 171 | NA | NA |
| No. of LTMs | 525 | 58 | NA |
| No. of HTMs | 8307 | 1772 | NA |
| No. of HTMs validated by low-throughput methoda | 283 | NA | NA |
| Cell lines/tissues | 373 | ∼25 | NA |
| Enrichment druga | 63 | NA | NA |
| Therapeutic target | 445 | NA | 135 |
| Drugs/small-molecule inhibitor | 383 | NA | 118 |
| Bioassay method | 6 | NA | 20 |
| Mentioned effect of the drug on CSCs traita | Yes | NA | NA |
| Attributes . | BCSCdb . | CSCdb . | CSCTT . |
|---|---|---|---|
| Validation of biomarkers in the clinical sample | 171 | NA | NA |
| No. of LTMs | 525 | 58 | NA |
| No. of HTMs | 8307 | 1772 | NA |
| No. of HTMs validated by low-throughput methoda | 283 | NA | NA |
| Cell lines/tissues | 373 | ∼25 | NA |
| Enrichment druga | 63 | NA | NA |
| Therapeutic target | 445 | NA | 135 |
| Drugs/small-molecule inhibitor | 383 | NA | 118 |
| Bioassay method | 6 | NA | 20 |
| Mentioned effect of the drug on CSCs traita | Yes | NA | NA |
NA = Not available.
Indicates the unique attributes of BCSCdb.
Comparison of attributes and data content of BCSCdb with CSCdb and CSCTT
| Attributes . | BCSCdb . | CSCdb . | CSCTT . |
|---|---|---|---|
| Validation of biomarkers in the clinical sample | 171 | NA | NA |
| No. of LTMs | 525 | 58 | NA |
| No. of HTMs | 8307 | 1772 | NA |
| No. of HTMs validated by low-throughput methoda | 283 | NA | NA |
| Cell lines/tissues | 373 | ∼25 | NA |
| Enrichment druga | 63 | NA | NA |
| Therapeutic target | 445 | NA | 135 |
| Drugs/small-molecule inhibitor | 383 | NA | 118 |
| Bioassay method | 6 | NA | 20 |
| Mentioned effect of the drug on CSCs traita | Yes | NA | NA |
| Attributes . | BCSCdb . | CSCdb . | CSCTT . |
|---|---|---|---|
| Validation of biomarkers in the clinical sample | 171 | NA | NA |
| No. of LTMs | 525 | 58 | NA |
| No. of HTMs | 8307 | 1772 | NA |
| No. of HTMs validated by low-throughput methoda | 283 | NA | NA |
| Cell lines/tissues | 373 | ∼25 | NA |
| Enrichment druga | 63 | NA | NA |
| Therapeutic target | 445 | NA | 135 |
| Drugs/small-molecule inhibitor | 383 | NA | 118 |
| Bioassay method | 6 | NA | 20 |
| Mentioned effect of the drug on CSCs traita | Yes | NA | NA |
NA = Not available.
Indicates the unique attributes of BCSCdb.
Discussion
BCSCdb is an important web portal for CSC research. It compiles human cell line-specific experimentally validated CSC-related low-throughput and high-throughput biomarkers information, CSC-related therapeutics and CSC biomarkers interaction data into a single platform. It provides methods that are in use for the identification of CSC populations. Users can also get to know about the experimental method that has been used to identify a specific CSC biomarker for a particular cancer cell line and clinical biomarker from primary tissue (25–30). It also provides a list of drugs that can enrich the CSC population such as tamoxifen, irinotecan and dasatinib (31–33). We focus on the CSC biomarker’s quality assessment by providing two scoring systems. A confidence score inculcates the confidence of a biomarker identification by one or more experimental methods in a particular CSC cell line. Moreover, the global scoring of CSC biomarkers can provide an idea about the unique and shared biomarkers. If a CSC biomarker has a higher global score, then the biomarker could be considered a true global marker for different types of cancer. Similarly, if a CSC biomarker has a low global score value, then it can denote the uniqueness of the CSC biomarker for that particular type of cancer.
The CSC biomarker molecular interaction data were obtained from specific cancer cell lines, and those interactions were experimentally validated in CSC populations. Five different types of interaction data from different types of CSC populations including (i) protein–protein interaction, (ii) protein–gene interaction, (iii) gene–gene interaction, (iv) miRNA–mRNA interaction and (v) RNA–protein interaction were reported. There are a few limitations to the study: the gene–gene interaction data for CSCs are not available on the browse page of BCSCdb in the Cytoscape network. Therefore, in the interaction network model, we have only included the protein–protein, gene–protein, miRNA–mRNA and RNA–protein interactions in network formation. Among 373 cell lines, only 16 cell lines were reported as having more than five interactions, and we have considered only these cell lines for the Cytoscape network model. The cell lines for network formation included lung cancer, hepatic cancer, colon cancer, breast cancer, pancreatic cancer and gastric cancer. Another limitation of the study is, in therapeutics, single as well as combination drugs strategies have been used to target CSCs; however, we have kept a single PubChem ID for the unique drug in the table whenever two drugs were used at the same time to target the CSC population. The global score of each CSC biomarker is dependent on the total number of entries, so in each updated version, these values will change. This could be a limitation of this database. But the changes in global score will be in the hundredths or thousandths place (second or third digit to the right of the decimal point) since the denominator or total of PMID (PubMed IDs) will change.
We have selected a threshold value for confidence and global scores and identified the potential CSC biomarkers that can serve as a potential therapeutic target to eradicate the CSC population. The moderate threshold values give a hit of common 57 genes from both the scoring systems that have high global as well as a confidence score. Furthermore, a number of studies have demonstrated the role of miRNA in regulating the CSC’s self-renewal and differentiation, and for many CSCs, miRNA biomarkers have also been reported in different types of cancer (34). We have identified that MIR15B, MIR30A and MIR19A act as common high-throughput biomarkers for breast, lung and pancreatic cancer (35, 36). The expression of MIR15B has also been reported through low-throughput studies in hepatic cancer, and recent studies suggested that miR15B regulates metastasis in breast cancer and acts as a novel therapeutic target for esophageal cancer (37, 38). Thereby, among other reported miRNA biomarkers, MIR15B can be considered at a global level, and its expression in other CSC populations could be explored in different types of cancer, such as bladder cancer and head and neck cancer.
The BCSCdb is a user-friendly database and is designed in a way that the search and the advanced search option output will display the high-scoring CSC biomarkers at the top of the list and help researchers identify the novel CSC biomarker along with the detection method in a said cancer type. In our case study, we observed that DCLK1 can be a promising CSC biomarker for colon cancer since it has a very high confidence score specific for colon cancer and a low global score. We are expecting that BCSCdb will help advance the CSC research and facilitate the identification of novel CSC biomarkers.
Supplementary data
Supplementary data are available at Database Online.
Acknowledgements
S.F. and A.G. are thankful to the University Grant Commission and Department of Biotechnology, Ministry of Science and Technology, Government of India, for their respective fellowships. The authors are also thankful to the Division of Bioinformatics, Bose Institute, Kolkata, for providing the infrastructure to perform this work.
Funding
Bioinformatics Center (BIC at Bose Institute, Kolkata, to S.S.), funded by the Department of Biotechnology, Government of India, vide the sanction no. BT/PR40174/BTIS/137/45/2022. Funding for open access publication charges: Bose Institute, Kolkata, India.
Conflict of interest
The authors declare no competing financial interests.
Data availability
Prior registration or password is not required, and BCSCdb is easily accessible at http://dibresources.jcbose.ac.in/ssaha4/bcscdb.