-
PDF
- Split View
-
Views
-
Cite
Cite
Andrew Chatr-aryamontri, Lynette Hirschman, Karen E Ross, Rose Oughtred, Martin Krallinger, Kara Dolinski, Mike Tyers, Tonia Korves, Cecilia N Arighi, Overview of the COVID-19 text mining tool interactive demonstration track in BioCreative VII, Database, Volume 2022, 2022, baac084, https://doi.org/10.1093/database/baac084
Close - Share Icon Share
Abstract
The coronavirus disease 2019 (COVID-19) pandemic has compelled biomedical researchers to communicate data in real time to establish more effective medical treatments and public health policies. Nontraditional sources such as preprint publications, i.e. articles not yet validated by peer review, have become crucial hubs for the dissemination of scientific results. Natural language processing (NLP) systems have been recently developed to extract and organize COVID-19 data in reasoning systems. Given this scenario, the BioCreative COVID-19 text mining tool interactive demonstration track was created to assess the landscape of the available tools and to gauge user interest, thereby providing a two-way communication channel between NLP system developers and potential end users. The goal was to inform system designers about the performance and usability of their products and to suggest new additional features. Considering the exploratory nature of this track, the call for participation solicited teams to apply for the track, based on their system’s ability to perform COVID-19-related tasks and interest in receiving user feedback. We also recruited volunteer users to test systems. Seven teams registered systems for the track, and >30 individuals volunteered as test users; these volunteer users covered a broad range of specialties, including bench scientists, bioinformaticians and biocurators. The users, who had the option to participate anonymously, were provided with written and video documentation to familiarize themselves with the NLP tools and completed a survey to record their evaluation. Additional feedback was also provided by NLP system developers. The track was well received as shown by the overall positive feedback from the participating teams and the users.
Database URL: https://biocreative.bioinformatics.udel.edu/tasks/biocreative-vii/track-4/
Introduction
BioCreative (Critical Assessment of Information Extraction in Biology) is a community effort for the assessment of the state of the art in text mining and information extraction technologies applied to the biomedical domain (1). Since its inception in 2004, BioCreative has evaluated systems and algorithms for the completion of numerous tasks, including bioentity mention identification, normalization of biomedical entities (genes and compounds) (2–5) and the identification of functional relationships such as protein–protein or chemical–disease interactions (6, 7).
In light of the interdisciplinary scope of BioCreative, the interactive task (IAT) was first introduced in the third edition of BioCreative (8–10) to bring together experts from text mining and biocuration to support real-life tasks by providing component modules for text mining services. Initially, the IAT focused on the needs of the biocuration community, a natural user community for the BioCreative activities; this represented an opportunity to implement text mining algorithms and methods into prototypes that could be directly used and evaluated by experienced curators from specialized databases. Such an approach proved to be beneficial to both system developers and database curators. In particular, system developers benefited from the detailed feedback provided by the testers that allowed developers to improve and implement new functionalities, while curators were provided with improved tools for data curation.
The emergence of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus in late 2019 and the consequent global health crisis caused by the coronavirus disease 2019 (COVID-19) pandemic in early 2020 posed great challenges to biomedical researchers, policymakers and health officials with its poorly understood biology and complete absence of vaccines or effective therapeutics. To date, >570 million infections and >6 million deaths have been documented, although the actual numbers are likely to be far higher (https://covid19.who.int/ as of 1 August 2022). Researchers worldwide rushed to produce new data and analyses in order to elucidate the biology of SARS-CoV-2 and to identify potential treatments for COVID-19. Because it became crucial to exchange experimental results as rapidly as possible, one consequence of the crisis was a change in how experimental findings were disseminated. In a departure from the traditional route of reporting results in journals and conference proceedings after the time-consuming peer-review process, preprint publications not yet validated by rounds of peer reviewing became the primary means of disseminating results on SARS-CoV-2 and COVID-19. In 2020, >25% of the 125 000 COVID-19-related articles were preprints (11). In addition, the drive to identify COVID-19 therapeutics based on novel bioactive compounds and drug repurposing made other sources, such as clinical trials repositories (e.g., the World Health Organization (WHO) International Clinical Trials Registry Platform), a fundamental tool in the fight against the global pandemic. Unfortunately, these repositories were not adequately equipped to provide worldwide support and functionalities to researchers and physicians. Echoing the experimental efforts, the bioinformatics community contributed with a variety of tools either by customization of existing tools or via the development of new customized tools (12). Primary biomedical repositories, such as National Center for Biotechnology Information (NCBI) (13) and Uniprot (14), implemented accelerated genome and proteome annotation pipelines for SARS-CoV-2 strains. Other tools provided support to track and predict the epidemiology of the pandemic (15), study coronavirus evolution (16, 17) and facilitate drug design and discovery (18, 19).
Natural language processing (NLP) experts made available a plethora of tools to support not only researchers but also public health officials and policymakers in their strategic planning to contain the pandemic. Text mining approaches provided solutions covering a broad spectrum of applications from generating COVID-19 corpora (20, 21) in different languages to the conceptualization and realization of search engines leveraging retrieval extraction and classification algorithms. NLP modules were also instrumental in the development of reasoning tools and knowledge discovery systems (21).
The proliferation of text mining tools for COVID-19 applications motivated the launch of the COVID-19 text mining tool interactive demonstration track (demo track) in BioCreative VII. The track was analogous to IAT tracks held in past editions, but it was structured differently because the participating systems were not required to perform a specific task in a competitive format. While most open challenge evaluations involve the evaluation of systems on a specific task or tasks, this demo track was intended as a demonstration and collaborative task to formally assess the performance of selected systems to enhance SARS-CoV-2 and COVID-19 research. The participating systems targeted a variety of users, including researchers, clinicians, biocurators and policymakers, and offered solutions for preprint aggregation or leveraged the available literature to provide knowledge graph and reasoning systems. An appealing feature of the interactive demo track is the opportunity for the participating teams to establish two-way communication with the users testing the NLP systems and to receive a detailed report on system functionality and performance. Because NLP system developers rarely interact with their target audience during the instantiation of their systems, accurate and individualized feedback provides a unique opportunity to refine the user–system experience and to efficiently identify areas for further improvement. A collateral benefit of the demo track is to increase the visibility of NLP systems across a large and diverse audience, as well as exposing the user community to new NLP tools.
Task design and organization
The COVID-19 text mining interactive demo task was designed as an exploratory demonstration task for the assessment of web/application programming interface (API) or standalone interfaces with a text mining back-end devoted to supporting COVID-19 research. The call for participation went out at the end of February 2021, and candidate teams had to submit their application by 7 June 2021. Seven teams applied and all were accepted for participation (notification on 20 June). In order to participate, candidate teams were required to submit documentation detailing the objective of their system and the tasks it performs (e.g. information retrieval, relation extraction and topic clustering), the targeted users and the example use cases. In addition, registrants had to provide technical details, including the URL and web browser compatibility, details on the user interactivity options (highlighting, sorting, filtering, editing and exporting results), data sources and a system performance report if available. This documentation was needed because the task is centered around the human–machine interaction and trivial pitfalls of the interface would penalize the NLP module powering the tool. Once selected, teams were asked to provide additional documentation (by 23 July 2022) in the form of tutorials and instructional videos to illustrate the functionalities of each system and to provide use cases and examples. This material was made available on the BioCreative Track IV page (URL: https://biocreative.bioinformatics.udel.edu/tasks/biocreative-vii/track-4-users/) and was instrumental in the recruitment of test users.
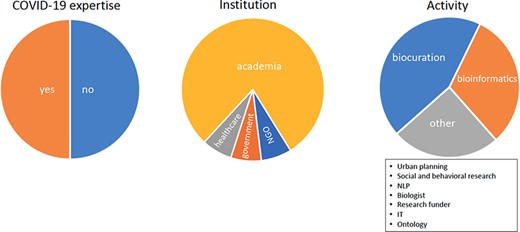
Because the systems involved were not required to perform any specific task, the user group was not strictly composed of biocurators, and teams were encouraged to provide contacts for potential testers to ensure that the appropriate target user community was represented in the group of users. In order to recruit the users, information about the track was disseminated via multiple avenues, including social media (Twitter and Facebook), mailing lists (e.g. societies, academic research departments and pharmaceutical companies) and by contacting research domain experts. Additional users were recruited from the staff of various biomedical databases. Most recruited users were from academia, with a significant fraction working in biocuration or bioinformatics. Approximately 50% of users worked in research related to COVID-19, and ∼75% of these were employed in academia (Figure 1). Individuals interested in participating as testers had access to a web page (https://biocreative.bioinformatics.udel.edu/tasks/biocreative-vii/track-4-users) providing a complete overview of the systems and information needed to complete the tasks (Figure 2). Upon choosing the system of interest, the users familiarized themselves with its features and capabilities by completing a guided activity prepared by each system. After that, they completed an exploratory phase where they were asked to independently navigate the systems and test their own example publications and/or preprints.
Pie charts show the COVID-19 expertise, the place of employment and the position roles of users.
Metrics of evaluation
After testing one or more NLP systems, users were asked to fill out an anonymous survey. The survey was divided into three different sections designed to gather information about the professional background of the user, assess the NLP system(s) and provide an overall evaluation of their user experience. The first section was designed to learn about the user’s involvement in COVID-19-related research, the type of institution where they were employed (e.g. academia, pharma and health care) and their professional role (e.g. biocurator, clinician and researcher). The second section was designed to collect user feedback and recommendations and was mainly based on a set of open-ended questions. The users rated their overall level of satisfaction with the system and provided their opinions on system functionalities, addressing usability and potential bottlenecks, organization of the output and the possibility of exporting the results in multiple formats. The users were also asked about their prior experience with similar systems and if they considered the system potentially useful for their work or that of others.
The last section contained a system usability score (SUS) (22), a post-test questionnaire with 10 questions answered via a Likert scale (1–5). The SUS is routinely used to assess the usability of a system by evaluating the ability of the user to successfully achieve the task, the effort needed to complete the task and user satisfaction (9). The odd-numbered questions asked the user to agree/disagree with some positive aspect (e.g. ‘I thought this system was easy to use’), whereas even-numbered ones were centered around negative features (e.g. ‘I found this system unnecessarily complex’). To calculate the SUS score, the score contribution for each item was normalized to a range of 0–4 (for positive (odd-numbered) questions, the score contribution was the scale position minus 1; for negative (even-numbered) questions, the contribution was 5 minus the scale position). The total score was then multiplied by 2.5 to convert a range from 0 to 100 , where higher scores indicated great usability.
In this third section, we also included two questions about the user’s overall impression of the NLP system and whether the system met expectations, to which users responded using a Likert scale ranging from 1 (worst) to 5 (best). For each system, we calculated the percentage of users assigning scores <3 (negative impression), >3 (positive impression) and = 3 (neutral impression).
In addition, participant teams answered a separate survey created to evaluate the reception of the track from the system developers’ perspective. Teams were asked to evaluate the usefulness of the users’ comments, if the feedback prompted any improvement in the system, and whether the number and the background of the reviewers were adequate. The questions were answered with a Likert scale (1–5) with the responses ranging from 1 (worst) to 5 (best), and scores <3, >3 or = 3 were considered negative, positive or neutral, respectively.
Participating teams
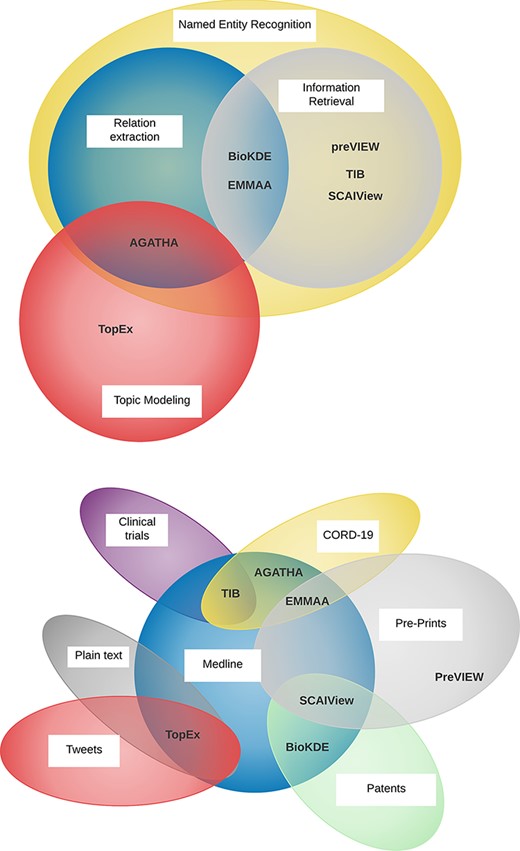
Seven NLP systems were applied to participate in this track. The participants varied in scope (information retrieval, named entity recognition, relation extraction and topic modeling), implementation (search engine, knowledge graph, hypothesis generation) and in the source of the input texts (abstracts, preprints, clinical trials and tweets) (Figure 3). All of the NLP systems targeted similar user communities with no specific NLP or informatics expertise: biomedical researchers, curators, translational researchers and clinicians with an interest in COVID-19. Some of the development teams also targeted users from government agencies. For example, the Therapeutic Information Browser (TIB) (23) team identified funding agency decision-makers working on COVID-19 therapeutics, and the COVID-19 SCAIView team identified organizations such as the WHO Pandemic Hub and the COVID-19 Data Portal.
Venn diagrams showing the diversity of the tasks performed by the participating NLP teams (top) and the various sources of textual data (bottom).
The TIB enables the user to easily find and track scientific information about potential COVID-19 therapeutics and vaccines. The system employs rule-based NLP to identify literature and clinical trials pertinent to drugs and selected viruses and provides additional details on the type of study where the drug was evaluated (e.g. cell-based, animal or clinical). preVIEW COVID-19 (24) performs searches over seven different preprint servers and provides the user with filtering options based on semantic concepts associated with SARS-CoV-2 variants and proteins, human genes and diseases. SCAIView (25) carries out semantic searches in large text collections, based on a combination of free-text and ontological representation of biomedical entities. The ontological terminology is highlighted on the result pages where additional filtering based on ontological concepts or publication features can be used to refine the results.
BioKDE (26) is a biomedical knowledge discovery platform consisting of two main modules, a search engine mining PubMed and a knowledge graph-based visualization system. The results can be visualized in a knowledge graph where detected entities are represented as nodes and edges represent their relationships.
EMMAA (27) and AGATHA (28) leverage COVID-19 data retrieved from the literature to create models and generate hypotheses to help researchers discover explicit or implicit connections between biomedical entities. EMMAA is a framework for extracting causal and mechanistic relations for COVID-19. EMMAA takes advantage of several available text mining tools to identify and extract the pertinent relations from the biomedical literature to create a knowledge graph. Relations from the knowledge graph can then be assembled into a self-updating model which users can query interactively. The model also supports causal path-based analysis to decipher the effect of drugs. AGATHA generates scientific hypotheses derived by the extraction and analysis of semantic concepts. The tool is adjustable via many parameters that provide the users with additional flexibility in their decision-making process.
TopEx (29) is a domain agnostic topic modeling tool that enables nontechnical users, via a user-friendly interface, to explore topics in a corpus of text documents. TopEx is designed to work with niche-corpora uploaded by the user. Key topics are identified by grouping semantically similar sentences and performing a topic analysis on each group. TopEx facilitates document classification and allows the user to visualize the evolution of trends in topics over time.
Survey results
Turning survey results into a clear analysis is not straightforward, and this was especially true in the case of Track IV due to the relatively small number of users and because each system was tested by a different group. In addition, each system used NLP in a somewhat different way. Nonetheless, the survey provided insights on important tool features from a user perspective and answers about which areas need more development. Through the free-text section of the survey, the users could provide their level of familiarity with similar systems (Figure 4A) and highlight the obstacles encountered with each system pertaining to lack of functionalities, quality of documentation, bugs and performance issues (Figure 4B).
The two bar plots display the familiarity of the users with systems similar to the ones participating in Track IV (A) and the most common bottlenecks encountered in the evaluation of the systems (B).
The word clouds in Figure 5 show the qualities most appreciated by the users for each system. As expected, easy-to-use and intuitive systems garnered the most positive feedback, especially if some level of interactivity was provided to the user. Graphics features that facilitate and enhance the utility of the results were also valued, as was the capability to customize the output page by applying additional filters or highlighting semantic or ontological concepts. The users also stressed the importance of ranked results wherever possible, as well as the option to download in multiple formats, and extensive and clear documentation. It is to be noted that, in light of the diversity of the systems, the users were not provided with a controlled vocabulary to describe the impressions/features of the systems, nor were the concepts normalized at a later stage.
Word cloud representation generated with aggregated user comments about their favorite system features. Comments identified unique aspects of the systems such as comprehensive sources for preprints in preVIEW, highlighting concepts in semantic search engines like SCAIView filters and data organization in TIB, graph capabilities and interactivity in BioKDE and EMMAA, topic paths in AGATHA and finding trends in TopEX.
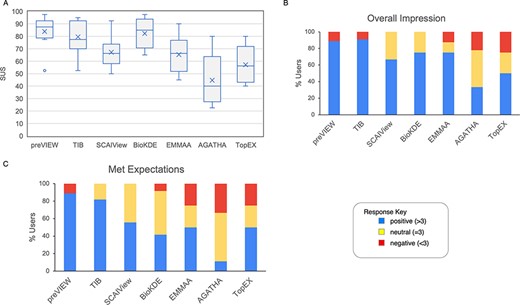
The analysis of the SUS confirmed a generally positive reception of the various systems among the users, as did the aggregate results for the ‘Overall impression’ and ‘Met expectations’ questions, as measured by the associated Likert scale (Figure 6). It should be noted that the lower rating of some tools, such as TopEX and AGATHA, was probably linked to the lack of familiarity of users with similar systems. On a positive note, many users found the tested NLP systems to be equally or more effective than systems they were already familiar with.
Results from questions evaluated with a Likert scale. (A) The boxplot represents the SUS distribution for each system. The X represents the mean, the horizontal line within the box is the median and the circle is an outlier. (B) Aggregate response to the ‘Overall impression’ for the systems. Scores >3, <3 and =3 were labeled as positive, negative and neutral impression, respectively. (C) Aggregate response to the question ‘meeting expectations’.
The developers confirmed that the feedback received from the users and the organizers prior to the task was instrumental in upgrading system functionalities and identifying areas of improvement. In particular, the exposure of the systems to testers with different types of expertise provided useful input for new options and applications. The various NLP teams expressed interest in a more iterative process of evaluation with multiple rounds of direct communication with the users. The NLP groups also lamented the limited size of the user group (six to eight individuals) and a lack of representation of their intended target audience, such as bench scientists and clinicians.
An additional benefit of the task was that NLP developers were compelled to create clear written and visual documentation from the perspective of a user. Unexpectedly, many NLP teams requested a more competitive evaluation in the form of a shared task and to introduce more objective metrics, such as search speed, in the user survey.
Discussion and future perspectives
The organization of this edition of the IAT posed many challenges. Most open challenge evaluations involve the evaluation of system performance on a specific task or tasks because this ensures comparability of the systems. However, the diversity of the tasks performed by the systems and the assortment of employed technologies, including the use of unlabeled data, made it impractical to make a direct comparison across all participants. Moreover, the NLP systems that participated in this task (e.g. topic modeling and knowledge graphs) did not represent the more traditional curation suites usually developed under BioCreative. This led us to an evaluation approach based on usability (30). This approach assesses the experiences and reactions of users exercising an ‘individual’ system for typical tasks. This approach has been used (31) to drive community progress toward systems that collaborate with humans on a variety of complex, open-ended activities. The usability-based approach made it possible to provide system-specific feedback to developers—a major goal of the BioCreative demo task.
The recruitment of a large and appropriate body of users was complicated by the fact that target users were mainly bench biologists, medical practitioners and pharma employees. In prior BioCreative interactive tracks, systems were required to perform biocuration tasks, making it relatively easy to gather a testing group from specialized databases. However, in this round, with tools designed mainly for practitioners, it was very difficult to assemble an appropriate testing cohort. Overall, the composition of the user group was considered sufficient in size and quality, although two NLP teams criticized the limited presence of target users. It is not clear whether the difficulty in recruiting volunteers reflects a lack of interest or requires identifying more efficient community engagement, other than biocurators who usually participate in BioCreative. Regardless, biocurators, who usually hold a PhD and have extensive experimental experience, work in a variety of settings and with various types of data (literature, clinical and genomics) and are therefore, in general, well equipped to review NLP systems.
The success of this task and the positive feedback from both the participating NLP teams and the users frames the discussion about future editions of this track. A more traditional organization of the track, with tasks focused mainly on biocuration, would facilitate the comparison and evaluation of the participating teams. On the other hand, a more open exploratory setup allows coverage of a broader range of applications and potentially appeals to a larger audience.
Is it possible to establish metrics to evaluate the success of the track by assessing the level of improvement of each system or gauging their penetration in the biomedical community? Can BioCreative contribute to the survival of NLP systems that are plagued by a lack of consistent funding support? It will be crucial to find a balance between all of these interests in order to derive the highest possible benefit from the interactive track, especially given that organization and participation in BioCreative are resource-intensive and time-consuming for all the parties involved. However, getting an equivalent set of evaluation results and user feedback would be much more difficult and time-consuming if conducted individually outside of an organization like BioCreative.
Acknowledgements
We would like to acknowledge the NLP teams who participated in Track IV and the users who took the time to review the systems and provide valuable feedback. We also would like to acknowledge the International Society for Biocuration (ISB) and the National Library of Medicine (NLM) for their dedicated efforts in disseminating our track and helping us recruit participants.
Funding
National Institutes of Health Office of Research Infrastructure Programs (R01OD010929 to M.T. and K.D.); Canadian Institutes of Health Research (FDN-167277 to M.T.); Canada Research Chair in Systems and Synthetic Biology (to M.T.); National Institutes of Health (2U24HG007822-08, 1R35 GM141873-01 to K.E.R. and C.N.A); Spanish Plan for the Advancement of Language Technology and Proyectos I+D+i2020-AI4PROFHEALTH (PID2020-119266RA-I00 to M.K.); MITRE (W56KGU-18-D-0004 to L.H. and T.K.). The views, opinions and/or findings contained in this report are those of the authors and should not be construed as an official government position, policy or decision.
Conflict of interest
None declared.