-
PDF
- Split View
-
Views
-
Cite
Cite
Jin Li, Jing Gao, Baiyang Feng, Yi Jing, PlagueKD: a knowledge graph–based plague knowledge database, Database, Volume 2022, 2022, baac100, https://doi.org/10.1093/database/baac100
Close - Share Icon Share
Abstract
Plague has been confirmed as an extremely horrific international quarantine infectious disease attributed to Yersinia pestis. It has an extraordinarily high lethal rate that poses a serious hazard to human and animal lives. With the deepening of research, there has been a considerable amount of literature related to the plague that has never been systematically integrated. Indeed, it makes researchers time-consuming and laborious when they conduct some investigation. Accordingly, integrating and excavating plague-related knowledge from considerable literature takes on a critical significance. Moreover, a comprehensive plague knowledge base should be urgently built. To solve the above issues, the plague knowledge base is built for the first time. A database is built from the literature mining based on knowledge graph, which is capable of storing, retrieving, managing and accessing data. First, 5388 plague-related abstracts that were obtained automatically from PubMed are integrated, and plague entity dictionary and ontology knowledge base are constructed by using text mining technology. Second, the scattered plague-related knowledge is correlated through knowledge graph technology. A multifactor correlation knowledge graph centered on plague is formed, which contains 9633 nodes of 33 types (e.g. disease, gene, protein, species, symptom, treatment and geographic location), as well as 9466 association relations (e.g. disease–gene, gene–protein and disease–species). The Neo4j graph database is adopted to store and manage the relational data in the form of triple. Lastly, a plague knowledge base is built, which can successfully manage and visualize a large amount of structured plague-related data. This knowledge base almost provides an integrated and comprehensive plague-related knowledge. It should not only help researchers to better understand the complex pathogenesis and potential therapeutic approaches of plague but also take on a key significance to reference for exploring potential action mechanisms of corresponding drug candidates and the development of vaccine in the future. Furthermore, it is of great significance to promote the field of plague research. Researchers are enabled to acquire data more easily for more effective research.
Database URL: http://39.104.28.169:18095/
Introduction
Plague, also known as the Black Death, is a global zoonotic disease caused by Yersinia pestis (1). It was once a Class A biological and chemical weapon in warfare. Its pathogenic bacterium Y. pestis has been listed as a Class A bioterrorism agent (2). It poses a great hazard to humans and animals for its strong infectivity and high mortality. The disease primarily comprises five principal forms, including bubonic, pneumonic, septicemic, meningeal and pharyngeal plague (3). It can be transmitted by flea bites, respiratory droplets, eating uncooked contaminated meat and contacting infected pets/livestock, thus resulting in skin ulceration, carbuncles and ulcers, along with pustules, spots, petechiae, bruising and gangrene. If treatment is not prompt, patients can die from heart failure and shock (4–6). Thus, the potential pathogenic genes, risk factors and precursors of plague infection should be further investigated to find effective prevention and treatment methods.
With the explosive growth of domain data, considerable plague-related knowledge is scattered across the literature. To conduct multivariate correlation analysis to determine the effect of different factors on plague, researchers have no choice but to retrieve and integrate information from different databases and then organize them manually. It is such a tedious and complicated manual task, and constructing numerous relation databases is also time-consuming. The built thematic base suffers from imperfect data information and cannot be updated in real time due to the limited human resources, thus causing a great inconvenience to those who study plague and affecting the progress of research. Accordingly, an integrated plague knowledge base is urgently required.
To solve the above problems, a comprehensive plague knowledge base is built by us. First, text mining technology is adopted to collect and organize the plague-related knowledge in the literature. Next, a semi-automatic approach is employed to extract entities and relations from the literature with manual annotations and review, so as to integrate plague-related information (e.g. genes, proteins, detection methods, treatments and geographic locations), which are implicit in the literature. On that basis, logical relations are built and a plague entity dictionary and ontology knowledge base are formed. Next, a plague-related knowledge graph is constructed through the Neo4j graph database. The Neo4j graph database is capable of describing the relations in a more intuitive and clearly graphical manner. PlagueKG is beneficial to understand this type of complex disease (e.g. exploring the precursors and sequelae of plague infection, therapeutic drugs and methods). PlagueKG covers 33 types of 9633 nodes, 292 types of 9466 relations as well as 9583 triples. Lastly, Web technology is employed to build a comprehensive plague knowledge database. To the best of our knowledge, this has been the first knowledge database of plague, which provides real and rich knowledge.
Related work
There are two mainstream ways to build database thus far. One way is based on sequencing data (7), and the other is based on literature mining. The Gene Ontology Annotation (8) and the Kyoto Encyclopedia of Genes and Genomes Orthology (KEGG) (9) are the most extensively used gene annotation databases. Gene Ontology (10) has been widely employed in the field of bioinformatics. KEGG is a comprehensive database integrating genomic, chemical and systemic functional information. Its metabolic pathway database refers to more authoritative public database. Other pathway databases, such as Reactome (11) and BioCarta (12), store protein annotation information, such as Swiss-Port (13) and Protein Data Bank (14), LncRNA and disease association database LncRNADisease (15). Some comprehensive databases, such as Entrez (16), BIND (17), biogrid (18), BGVD (19) and ChickVD (20), provide considerable information about genes and gene products, including pathway information and interaction information between genes.
To satisfy the needs of researchers to quickly gain insights into the current research status, the database based on literature mining has become a hotspot. Osteosarcoma–gene association database (21) is a literature mining database containing 911 genes and 81 microRNAs. PhamKG (22) is a multi-relational biomedical knowledge graph that includes over half a million entities of genes, drugs and diseases, and it builds relationships between each pair of entities. There are 29 types of relationship and more than 8000 entity vocabularies. To gain more insights into the genetic effect on human diseases, Phenomodifier, a manually managed database, provides a more complete spectrum of genetic factors contributing to human phenotypic variation. The database covers a total of 3078 modification information records, involving 288 different diseases, 2126 genetic modification variants as well as 843 different modification genes (23). Text-mined Hypertension, Obesity and Diabetes candidate gene database (T-HOD) (24) is another database regarding three common diseases. RAvariome is a database of genetic risk variants for rheumatoid arthritis (25).
Although the knowledge base built by the expert method has high accuracy, knowledge coverage is not comprehensive and not updated in a timely manner. The main reason is that the knowledge base has limited domain coverage due to the time-consuming and laborious process of manual review and editing, and the data update cannot keep abreast with the growth rate of newly published literature. Moreover, many biomedical fields still lack corresponding gene annotation library resources, and the gene information related to the above fields is scattered across thousands of literatures and not systematically collected.
Knowledge graph technology is capable of extracting structured knowledge from massive amounts of texts, which provides convenience for the extraction and display of literature knowledge (26). A knowledge graph is essentially a complex network that reflects the correlations between entities. It comprises nodes and edges capable of formally describing the real world and its relations. The nodes in a biomedical knowledge graph represent a variety of biological entities (e.g. genes, transcripts, proteins, pathways, diseases, symptoms, drugs and side effects), while the edges represent the logical or biological relations between entities (e.g. interaction, regulation, inhibition and inclusion.) (27, 28).
However, to the best of our knowledge, there has not been any research on the construction of the plague knowledge base yet. The lack of bioinformation ontology library for plague has significantly affected the automation of constructing the knowledge graph and knowledge base of plague and has reduced the accuracy of entity identification and relation extraction. The plague knowledge is scattered across a massive of literature, and it has not been collected and classified systematically. On that basis, it is urgently necessary to extract information and build a plague knowledge graph and knowledge base.
Methods
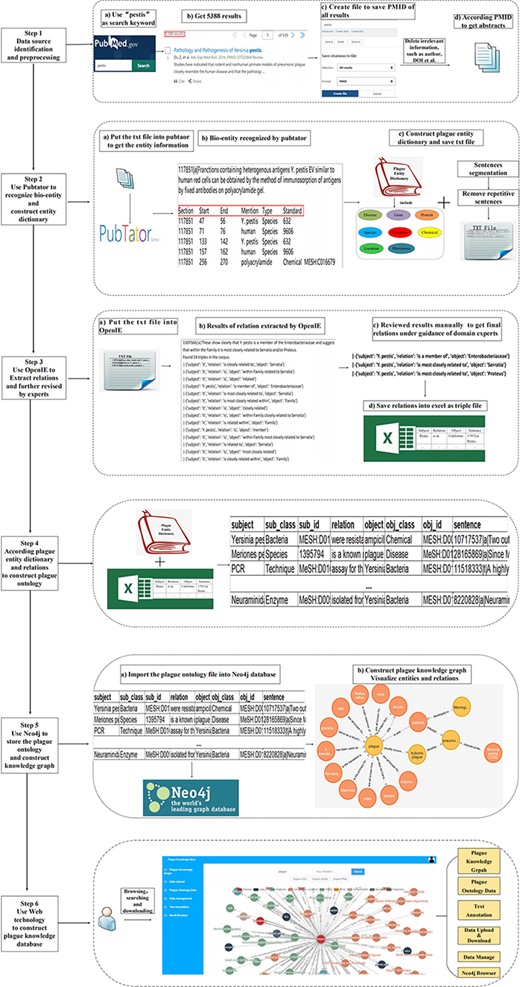
In this study, a workflow is designed to construct plague knowledge database from literature mining, identify plague-related entities and extract the relevant relations. The obtained literature abstracts are divided into 26 695 independent sentences, 33 kinds of the mentioned entity types are marked (i.e. gene, protein, drug, treatment, species and geographical location) and the plague entity dictionary is constructed. Subsequently, several methods are adopted to process the relations among entities, and plague ontology base is formed. Lastly, the plague knowledge graph and database are built respectively. The workflow is illustrated in Figure 1.
Materials and tools
The plague knowledge graph is built using a semi-automatic and semi-manual method. First, the plague-related abstracts are obtained from PubMed as a data source. Second, PubTator is adopted to recognize the entities in the abstracts. Next, the relations are extracted using OpenIE. Lastly, Neo4j is employed to store the information. In this part, the datasets and tools used in this study are briefed.
PubMed
PubMed is a free literature search engine. Although it has limitations on the access to full text, the abstracts have involved research purposes, methods, results and conclusions, basically covering the main part of information. In this study, we obtain relevant abstracts automatically, and use natural language processing techniques for text preprocessing. Meanwhile, we remove some irrelevant data such as author and publisher information and only retain the necessary information, as PubMed Unique Identifier (PMID), title and abstract, for subsequent knowledge extraction work.
PubTator
PubTator Central (29) is a Web-based system that can be accessed interactively through a Web browser. PubTator is employed as an entity recognition tool for the following three reasons: (i) PubTator only requires article’s PMID number to get the title and abstract; (ii) PubTator integrates multiple named entity recognition tools and can recognize six types of entities, such as GeneTUKit for gene extraction (30), tmVar for variant (31), DNorm for disease (32), dictionary-based method for chemicals and SR4GN for species (33) and (iii) the identified entities are marked with unique National Center for Biotechnology Information (NCBI_ID), such that further detailed information can be easily acquired.
OpenIE
In this study, we use the OpenIE tool for extracting relations semi-automatically. OpenIE is a relation extraction tool that extracts structured triples from text without specifying relations in advance (34).
Neo4j
Neo4j is a native graph database engine with a unique storage structure, index-free neighbor node storage method and a corresponding graph traversal algorithm, which makes retrieval speed faster. Its performance will not be affected with the increase in data, such that it exhibits significantly high query performance. Neo4j is more extensible and flexible than other graph databases. As an open-source database, its community version attracts numerous third parties’ utilization and promotion (35).
Data source
PubMed database is employed as the data source to build plague knowledge database. The literature is searched from PubMed with ‘pestis’ as the keyword, and the PMID numbers of all relevant research are obtained. Then, totally, 5388 corresponding article abstracts are obtained according to the PMID number automatically and stored in .txt format. To enrich the content of the knowledge base, all aliases of entities in the database of NCBI are obtained as to facilitate the retrieval in the knowledge graph at a later stage.
Data preprocessing
Since the obtained literature by PubTator covers some information that is not associated with the research (i.e. article publication time, author information and doi), abstracts are first preprocessed by a script to remove them and we make a clause operation, so as to facilitate subsequent work of named entity identification and relation extraction.
Named entity recognition
Thirty-three types of entities were recognized using a combination of PubTator, dictionary- and rule-based methods and manual annotation methods. PubTator is only capable of recognizing six types of entities (including gene, disease, chemical, mutation, species and cell line). On that basis, 27 entity types are added on the top of the plague knowledge database to expand the coverage of biomedical concepts and make this database more detailed and comprehensive. The following nine entity types (including amino acid, peptide, protein, lipid, enzyme, nucleotide, nucleic acid, toxin and vaccine) are further expanded in accordance with the chemical entity type provided by PubTator and Medical Subject Headings (MeSH), whereas the remaining 18 entity types (i.e. viruses, phenomenon, technique, anatomy, diagnosis, geographic location, symptom, environment, social sciences, etiology, assay, genome, equipment, therapeutics, time, person, single-nucleotide variant (SNV) and food) are constructed manually and individually in accordance with the text content and actual needs.
A plague entity dictionary is constructed based on the obtained entities and their types. In the process of constructing the dictionary, there are cases where the same entity corresponds to multiple different entry terms, so it is necessary to uniquely identify an entity based on the NCBI_ID. MeSH is the most frequently accessed database when we perform entity queries. It is an authoritative hierarchical subject term list compiled by the National Library of Medicine for indexing research in PubMed. In the process of retrieving entity categories, we select the most appropriate subject terms as the entity type based on the hierarchical relationship of the MeSH thesaurus, following the principle that the subject terms have high citation rates and match the actual text content information. On that basis, the unique entity mesh number is given. Since there are some errors when using PubTator to identify entities, such as classifying ‘California’ as a disease type, the unidentifiable entities should be annotated manually, and the annotated entities and their NCBI_ID should be corrected.
The samples of resultant data acquired after the named entity recognition are listed in Table 1. It covers PMID, starting position, end position, entity, type as well as attribute.
The sample result data of named entity recognition
| PMID . | Starting position . | End position . | Entity . | Type . | Attribute . |
|---|---|---|---|---|---|
| 896 271 | 461 | 465 | Mice | Species | 10 095 |
| 235 822 | 971 | 980 | Tularemia | Disease | MESH:D014406 |
| 896 271 | 461 | 465 | Guinea pigs | Species | 10 140 |
| 1 472 717 | 1016 | 1020 | LcrD | Protein | MeSH:C071579 |
| 848 916 | 229 | 245 | Benzylpenicillin | Chemical | MeSH:D010400 |
| PMID . | Starting position . | End position . | Entity . | Type . | Attribute . |
|---|---|---|---|---|---|
| 896 271 | 461 | 465 | Mice | Species | 10 095 |
| 235 822 | 971 | 980 | Tularemia | Disease | MESH:D014406 |
| 896 271 | 461 | 465 | Guinea pigs | Species | 10 140 |
| 1 472 717 | 1016 | 1020 | LcrD | Protein | MeSH:C071579 |
| 848 916 | 229 | 245 | Benzylpenicillin | Chemical | MeSH:D010400 |
The sample result data of named entity recognition
| PMID . | Starting position . | End position . | Entity . | Type . | Attribute . |
|---|---|---|---|---|---|
| 896 271 | 461 | 465 | Mice | Species | 10 095 |
| 235 822 | 971 | 980 | Tularemia | Disease | MESH:D014406 |
| 896 271 | 461 | 465 | Guinea pigs | Species | 10 140 |
| 1 472 717 | 1016 | 1020 | LcrD | Protein | MeSH:C071579 |
| 848 916 | 229 | 245 | Benzylpenicillin | Chemical | MeSH:D010400 |
| PMID . | Starting position . | End position . | Entity . | Type . | Attribute . |
|---|---|---|---|---|---|
| 896 271 | 461 | 465 | Mice | Species | 10 095 |
| 235 822 | 971 | 980 | Tularemia | Disease | MESH:D014406 |
| 896 271 | 461 | 465 | Guinea pigs | Species | 10 140 |
| 1 472 717 | 1016 | 1020 | LcrD | Protein | MeSH:C071579 |
| 848 916 | 229 | 245 | Benzylpenicillin | Chemical | MeSH:D010400 |
Table 2 lists the final constructed entity dictionary, which includes article PMID, entity name, type and NCBI_ID.
The sample of plague entity dictionary
| PMID . | Entity name . | Entity type . | NCBI_ID . |
|---|---|---|---|
| 117 851 | Human | Species | 9606 |
| 200 563 | Mouse | Species | 10 090 |
| 200 563 | Phenol | Chemical | MeSH:D019800 |
| 24 786 165 | Lymphadenopathy | Disease | MeSH:D000072281 |
| 24 786 165 | Vomiting | Symptom | MeSH:D014839 |
| 23 588 087 | IL-10 | Peptide | MeSH:D016753 |
| 1 695 896 | Insertion mutagenesis | Phenomena | MeSH:D016254 |
| 32 315 702 | JC221 | Viruses | 2 654 973 |
| 21 712 421 | Alpha-D-galactose | Enzyme | MeSH:D000519 |
| 21 219 468 | c-di-Cyclic diguanosine monophosphate (GMP) signaling | Nucleotide | MeSH:C062025 |
| 31 695 901 | Colorado | Geographic location | MeSH:D003120 |
| PMID . | Entity name . | Entity type . | NCBI_ID . |
|---|---|---|---|
| 117 851 | Human | Species | 9606 |
| 200 563 | Mouse | Species | 10 090 |
| 200 563 | Phenol | Chemical | MeSH:D019800 |
| 24 786 165 | Lymphadenopathy | Disease | MeSH:D000072281 |
| 24 786 165 | Vomiting | Symptom | MeSH:D014839 |
| 23 588 087 | IL-10 | Peptide | MeSH:D016753 |
| 1 695 896 | Insertion mutagenesis | Phenomena | MeSH:D016254 |
| 32 315 702 | JC221 | Viruses | 2 654 973 |
| 21 712 421 | Alpha-D-galactose | Enzyme | MeSH:D000519 |
| 21 219 468 | c-di-Cyclic diguanosine monophosphate (GMP) signaling | Nucleotide | MeSH:C062025 |
| 31 695 901 | Colorado | Geographic location | MeSH:D003120 |
The sample of plague entity dictionary
| PMID . | Entity name . | Entity type . | NCBI_ID . |
|---|---|---|---|
| 117 851 | Human | Species | 9606 |
| 200 563 | Mouse | Species | 10 090 |
| 200 563 | Phenol | Chemical | MeSH:D019800 |
| 24 786 165 | Lymphadenopathy | Disease | MeSH:D000072281 |
| 24 786 165 | Vomiting | Symptom | MeSH:D014839 |
| 23 588 087 | IL-10 | Peptide | MeSH:D016753 |
| 1 695 896 | Insertion mutagenesis | Phenomena | MeSH:D016254 |
| 32 315 702 | JC221 | Viruses | 2 654 973 |
| 21 712 421 | Alpha-D-galactose | Enzyme | MeSH:D000519 |
| 21 219 468 | c-di-Cyclic diguanosine monophosphate (GMP) signaling | Nucleotide | MeSH:C062025 |
| 31 695 901 | Colorado | Geographic location | MeSH:D003120 |
| PMID . | Entity name . | Entity type . | NCBI_ID . |
|---|---|---|---|
| 117 851 | Human | Species | 9606 |
| 200 563 | Mouse | Species | 10 090 |
| 200 563 | Phenol | Chemical | MeSH:D019800 |
| 24 786 165 | Lymphadenopathy | Disease | MeSH:D000072281 |
| 24 786 165 | Vomiting | Symptom | MeSH:D014839 |
| 23 588 087 | IL-10 | Peptide | MeSH:D016753 |
| 1 695 896 | Insertion mutagenesis | Phenomena | MeSH:D016254 |
| 32 315 702 | JC221 | Viruses | 2 654 973 |
| 21 712 421 | Alpha-D-galactose | Enzyme | MeSH:D000519 |
| 21 219 468 | c-di-Cyclic diguanosine monophosphate (GMP) signaling | Nucleotide | MeSH:C062025 |
| 31 695 901 | Colorado | Geographic location | MeSH:D003120 |
We list all the 33 kinds of entities and their several corresponding samples (Table 3).
The 33 kinds of entities
| S. No. . | Entity type . | Some samples of corresponding entities . |
|---|---|---|
| 1 | Gene | pst, pla, lcrH, etc. |
| 2 | Disease | Plague, lymphadenopathy, pneumonia, etc. |
| 3 | Chemical | Rifampicin, cefoperazone, cefotaxime, etc. |
| 4 | Species | Black rat, Oropsylla montana, guinea pigs, etc. |
| 5 | Cell | HeLa, SH-SY5Y, HEp-2, etc. |
| 6 | Protein | YopH, PrgI, MyfE, etc. |
| 7 | Bacteria | Y. pestis, Y. pseudotuberculosis, Escherichia coli, etc. |
| 8 | Amino acid | Citrulline, glycine, histidine, etc. |
| 9 | Peptide | Cytokines, interleukin-2, D-alanyl-D-alanine, etc. |
| 10 | Lipid | Lipid A, lipopolysaccharide, thromboxane, etc. |
| 11 | Enzyme | Alpha-D-galactose, beta-D-galactose, riboflavin, etc. |
| 12 | Nucleotide | c-di-Cyclic diguanosine monophosphate (GMP) signaling |
| 13 | Nucleic acid | Deoxyribonucleic acid, CsrB, 5S ribosomal RNAs, etc. |
| 14 | Toxin | Staphylococcal enterotoxin B, botulinum toxin |
| 15 | Vaccine | Live plague vaccine, rF1-V + alhydrogel, rF1 + rV, etc. |
| 16 | Viruses | Orthopoxviruses, influenza H7N9 viruses, arenaviruses, etc. |
| 17 | Phenomenon | Quorum sensing, phagocytosis, heat shock stimulons, etc. |
| 18 | Technique | loop-mediated isothermal amplification, polymerase chain reaction, enzyme immunoassay, etc. |
| 19 | Equipment | Microspheres, cryo electron microscopy, needles, etc. |
| 20 | Anatomy | Nostrils, intestinal epithelium, spleen, etc. |
| 21 | Diagnosis | Reaction of hemagglutination, Etest, western blotting, etc. |
| 22 | Geographic location | Vietnam, India, New Mexico, etc. |
| 23 | Symptom | Hypoxemia, difficulty breathing, myalgia, etc. |
| 24 | Environment | Mild winters, cool moist springs, temperate rainforest, etc. |
| 25 | Social Sciences | Bioterrorism |
| 26 | Etiology | Pathogenesis of plague, complications, transmission, etc. |
| 27 | Assay | N-Hydroxybenzimidazoles, ‘Bio T’ DNA assay |
| 28 | Genome | YAU, Y. pestis_A-1486, ASM252954v1, etc. |
| 29 | Therapeutics | Immunization, immunomodulation, chemoprophylaxis, etc. |
| 30 | Time | 8 years, 24 h, 3–4 days, etc. |
| 31 | Person | Pet ownership, cattle and sheep herdsmen, adult men, etc. |
| 32 | SNV | rs5030880 |
| 33 | Food | Raw ground beef, seafood, bottled water, etc. |
| S. No. . | Entity type . | Some samples of corresponding entities . |
|---|---|---|
| 1 | Gene | pst, pla, lcrH, etc. |
| 2 | Disease | Plague, lymphadenopathy, pneumonia, etc. |
| 3 | Chemical | Rifampicin, cefoperazone, cefotaxime, etc. |
| 4 | Species | Black rat, Oropsylla montana, guinea pigs, etc. |
| 5 | Cell | HeLa, SH-SY5Y, HEp-2, etc. |
| 6 | Protein | YopH, PrgI, MyfE, etc. |
| 7 | Bacteria | Y. pestis, Y. pseudotuberculosis, Escherichia coli, etc. |
| 8 | Amino acid | Citrulline, glycine, histidine, etc. |
| 9 | Peptide | Cytokines, interleukin-2, D-alanyl-D-alanine, etc. |
| 10 | Lipid | Lipid A, lipopolysaccharide, thromboxane, etc. |
| 11 | Enzyme | Alpha-D-galactose, beta-D-galactose, riboflavin, etc. |
| 12 | Nucleotide | c-di-Cyclic diguanosine monophosphate (GMP) signaling |
| 13 | Nucleic acid | Deoxyribonucleic acid, CsrB, 5S ribosomal RNAs, etc. |
| 14 | Toxin | Staphylococcal enterotoxin B, botulinum toxin |
| 15 | Vaccine | Live plague vaccine, rF1-V + alhydrogel, rF1 + rV, etc. |
| 16 | Viruses | Orthopoxviruses, influenza H7N9 viruses, arenaviruses, etc. |
| 17 | Phenomenon | Quorum sensing, phagocytosis, heat shock stimulons, etc. |
| 18 | Technique | loop-mediated isothermal amplification, polymerase chain reaction, enzyme immunoassay, etc. |
| 19 | Equipment | Microspheres, cryo electron microscopy, needles, etc. |
| 20 | Anatomy | Nostrils, intestinal epithelium, spleen, etc. |
| 21 | Diagnosis | Reaction of hemagglutination, Etest, western blotting, etc. |
| 22 | Geographic location | Vietnam, India, New Mexico, etc. |
| 23 | Symptom | Hypoxemia, difficulty breathing, myalgia, etc. |
| 24 | Environment | Mild winters, cool moist springs, temperate rainforest, etc. |
| 25 | Social Sciences | Bioterrorism |
| 26 | Etiology | Pathogenesis of plague, complications, transmission, etc. |
| 27 | Assay | N-Hydroxybenzimidazoles, ‘Bio T’ DNA assay |
| 28 | Genome | YAU, Y. pestis_A-1486, ASM252954v1, etc. |
| 29 | Therapeutics | Immunization, immunomodulation, chemoprophylaxis, etc. |
| 30 | Time | 8 years, 24 h, 3–4 days, etc. |
| 31 | Person | Pet ownership, cattle and sheep herdsmen, adult men, etc. |
| 32 | SNV | rs5030880 |
| 33 | Food | Raw ground beef, seafood, bottled water, etc. |
The 33 kinds of entities
| S. No. . | Entity type . | Some samples of corresponding entities . |
|---|---|---|
| 1 | Gene | pst, pla, lcrH, etc. |
| 2 | Disease | Plague, lymphadenopathy, pneumonia, etc. |
| 3 | Chemical | Rifampicin, cefoperazone, cefotaxime, etc. |
| 4 | Species | Black rat, Oropsylla montana, guinea pigs, etc. |
| 5 | Cell | HeLa, SH-SY5Y, HEp-2, etc. |
| 6 | Protein | YopH, PrgI, MyfE, etc. |
| 7 | Bacteria | Y. pestis, Y. pseudotuberculosis, Escherichia coli, etc. |
| 8 | Amino acid | Citrulline, glycine, histidine, etc. |
| 9 | Peptide | Cytokines, interleukin-2, D-alanyl-D-alanine, etc. |
| 10 | Lipid | Lipid A, lipopolysaccharide, thromboxane, etc. |
| 11 | Enzyme | Alpha-D-galactose, beta-D-galactose, riboflavin, etc. |
| 12 | Nucleotide | c-di-Cyclic diguanosine monophosphate (GMP) signaling |
| 13 | Nucleic acid | Deoxyribonucleic acid, CsrB, 5S ribosomal RNAs, etc. |
| 14 | Toxin | Staphylococcal enterotoxin B, botulinum toxin |
| 15 | Vaccine | Live plague vaccine, rF1-V + alhydrogel, rF1 + rV, etc. |
| 16 | Viruses | Orthopoxviruses, influenza H7N9 viruses, arenaviruses, etc. |
| 17 | Phenomenon | Quorum sensing, phagocytosis, heat shock stimulons, etc. |
| 18 | Technique | loop-mediated isothermal amplification, polymerase chain reaction, enzyme immunoassay, etc. |
| 19 | Equipment | Microspheres, cryo electron microscopy, needles, etc. |
| 20 | Anatomy | Nostrils, intestinal epithelium, spleen, etc. |
| 21 | Diagnosis | Reaction of hemagglutination, Etest, western blotting, etc. |
| 22 | Geographic location | Vietnam, India, New Mexico, etc. |
| 23 | Symptom | Hypoxemia, difficulty breathing, myalgia, etc. |
| 24 | Environment | Mild winters, cool moist springs, temperate rainforest, etc. |
| 25 | Social Sciences | Bioterrorism |
| 26 | Etiology | Pathogenesis of plague, complications, transmission, etc. |
| 27 | Assay | N-Hydroxybenzimidazoles, ‘Bio T’ DNA assay |
| 28 | Genome | YAU, Y. pestis_A-1486, ASM252954v1, etc. |
| 29 | Therapeutics | Immunization, immunomodulation, chemoprophylaxis, etc. |
| 30 | Time | 8 years, 24 h, 3–4 days, etc. |
| 31 | Person | Pet ownership, cattle and sheep herdsmen, adult men, etc. |
| 32 | SNV | rs5030880 |
| 33 | Food | Raw ground beef, seafood, bottled water, etc. |
| S. No. . | Entity type . | Some samples of corresponding entities . |
|---|---|---|
| 1 | Gene | pst, pla, lcrH, etc. |
| 2 | Disease | Plague, lymphadenopathy, pneumonia, etc. |
| 3 | Chemical | Rifampicin, cefoperazone, cefotaxime, etc. |
| 4 | Species | Black rat, Oropsylla montana, guinea pigs, etc. |
| 5 | Cell | HeLa, SH-SY5Y, HEp-2, etc. |
| 6 | Protein | YopH, PrgI, MyfE, etc. |
| 7 | Bacteria | Y. pestis, Y. pseudotuberculosis, Escherichia coli, etc. |
| 8 | Amino acid | Citrulline, glycine, histidine, etc. |
| 9 | Peptide | Cytokines, interleukin-2, D-alanyl-D-alanine, etc. |
| 10 | Lipid | Lipid A, lipopolysaccharide, thromboxane, etc. |
| 11 | Enzyme | Alpha-D-galactose, beta-D-galactose, riboflavin, etc. |
| 12 | Nucleotide | c-di-Cyclic diguanosine monophosphate (GMP) signaling |
| 13 | Nucleic acid | Deoxyribonucleic acid, CsrB, 5S ribosomal RNAs, etc. |
| 14 | Toxin | Staphylococcal enterotoxin B, botulinum toxin |
| 15 | Vaccine | Live plague vaccine, rF1-V + alhydrogel, rF1 + rV, etc. |
| 16 | Viruses | Orthopoxviruses, influenza H7N9 viruses, arenaviruses, etc. |
| 17 | Phenomenon | Quorum sensing, phagocytosis, heat shock stimulons, etc. |
| 18 | Technique | loop-mediated isothermal amplification, polymerase chain reaction, enzyme immunoassay, etc. |
| 19 | Equipment | Microspheres, cryo electron microscopy, needles, etc. |
| 20 | Anatomy | Nostrils, intestinal epithelium, spleen, etc. |
| 21 | Diagnosis | Reaction of hemagglutination, Etest, western blotting, etc. |
| 22 | Geographic location | Vietnam, India, New Mexico, etc. |
| 23 | Symptom | Hypoxemia, difficulty breathing, myalgia, etc. |
| 24 | Environment | Mild winters, cool moist springs, temperate rainforest, etc. |
| 25 | Social Sciences | Bioterrorism |
| 26 | Etiology | Pathogenesis of plague, complications, transmission, etc. |
| 27 | Assay | N-Hydroxybenzimidazoles, ‘Bio T’ DNA assay |
| 28 | Genome | YAU, Y. pestis_A-1486, ASM252954v1, etc. |
| 29 | Therapeutics | Immunization, immunomodulation, chemoprophylaxis, etc. |
| 30 | Time | 8 years, 24 h, 3–4 days, etc. |
| 31 | Person | Pet ownership, cattle and sheep herdsmen, adult men, etc. |
| 32 | SNV | rs5030880 |
| 33 | Food | Raw ground beef, seafood, bottled water, etc. |
Relation extraction
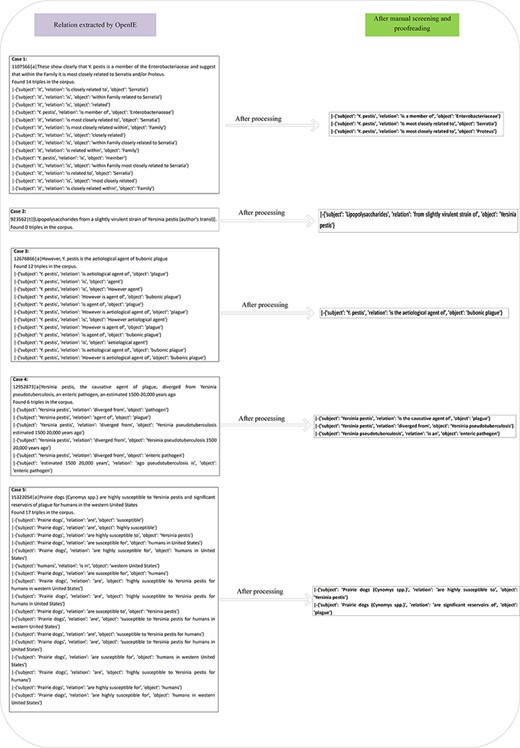
Abstracts are first divided into sentences by scripts. Subsequently, the sentences are de-duplicated. Lastly, a total of 26 695 sentences are obtained. Stanford OpenIE is employed to extract the entity relations. After extracting by OpenIE, the output results take the form of triple as {‘subject:’, ‘relationship:’, ‘object:’}. Since this study belongs to the vertical domain knowledge category, the relations are more complex and should be extracted in more details, as compared with the open domain. Besides, OpenIE is a general domain relation extraction tool, and its extraction results will be presented in a multi-possibility manner. On that basis, further manual screening and proofreading are required under the guidance of domain experts. As depicted in Figure 2, the left depicts the triples obtained by OpenIE and the right presents the triples obtained after manual review and proofreading by domain experts. For Case 1, 14 triples are extracted using OpenIE. Notably, the above triples do not clearly express the substantive and useful information. After processing, it yields three useful pieces of information. For Case 2, the sentence ‘923562|t|[Lipopolysaccharides from a slightly virulent strain of Yersinia pestis (author’s transl)]’, does have a relation, whereas OpenIE fails to extract the relationship. After manual curation by experts, it yields the triple ‘|-{“subject”: “Lipopolysaccharides”, “relation”: “from a slightly virulent strain of”, “object”: “Yersinia pestis”}’. For Case 3, only one triple is valid, so we remove the redundant ones. For Case 4, since the information extracted by OpenIE fails to accurately express the complete information of the sentence, after cleaning and manual curation by domain experts, we obtain three useful triples. For Case 5, after the manual review, we eventually get three triples. The above cases suggest that the data acquired after the manual review have higher quality and the semantic information is more accurate and complete.
Lastly, 9583 relations are obtained as gene—disease, disease—drug, disease—bacteria gene—protein, disease—species, etc. Tables 4 and 5 list the samples of detailed relation information and plague ontology data, respectively.
Samples of detailed relation information
| Subject . | Relation . | Object . | Sentence . |
|---|---|---|---|
| Biofilm formation | Critical for transmission | Y. pestis | 30 333 962|a|Biofilm formation is critical for blocking flea foregut and hence for transmission of Y. pestis by flea biting. |
| HmsA | InhibitION | Y. pestis virulence | 33 026 884|a|Conclusion: HmsA inhibits Y. pestis virulence, but this effect may be mediated by indirect effects on pathogenesis, iron homeostasis and/or other cellular processes. |
| LcrH | necessary for the normal response of Y. pestis | Adenosine triphosphate | 2 707 857|a|These findings show that LcrH is necessary for the normal response of Y. pestis to adenosine triphosphate and that LcrH contributes to Ca2+ respsonsiveness |
| Subject . | Relation . | Object . | Sentence . |
|---|---|---|---|
| Biofilm formation | Critical for transmission | Y. pestis | 30 333 962|a|Biofilm formation is critical for blocking flea foregut and hence for transmission of Y. pestis by flea biting. |
| HmsA | InhibitION | Y. pestis virulence | 33 026 884|a|Conclusion: HmsA inhibits Y. pestis virulence, but this effect may be mediated by indirect effects on pathogenesis, iron homeostasis and/or other cellular processes. |
| LcrH | necessary for the normal response of Y. pestis | Adenosine triphosphate | 2 707 857|a|These findings show that LcrH is necessary for the normal response of Y. pestis to adenosine triphosphate and that LcrH contributes to Ca2+ respsonsiveness |
Samples of detailed relation information
| Subject . | Relation . | Object . | Sentence . |
|---|---|---|---|
| Biofilm formation | Critical for transmission | Y. pestis | 30 333 962|a|Biofilm formation is critical for blocking flea foregut and hence for transmission of Y. pestis by flea biting. |
| HmsA | InhibitION | Y. pestis virulence | 33 026 884|a|Conclusion: HmsA inhibits Y. pestis virulence, but this effect may be mediated by indirect effects on pathogenesis, iron homeostasis and/or other cellular processes. |
| LcrH | necessary for the normal response of Y. pestis | Adenosine triphosphate | 2 707 857|a|These findings show that LcrH is necessary for the normal response of Y. pestis to adenosine triphosphate and that LcrH contributes to Ca2+ respsonsiveness |
| Subject . | Relation . | Object . | Sentence . |
|---|---|---|---|
| Biofilm formation | Critical for transmission | Y. pestis | 30 333 962|a|Biofilm formation is critical for blocking flea foregut and hence for transmission of Y. pestis by flea biting. |
| HmsA | InhibitION | Y. pestis virulence | 33 026 884|a|Conclusion: HmsA inhibits Y. pestis virulence, but this effect may be mediated by indirect effects on pathogenesis, iron homeostasis and/or other cellular processes. |
| LcrH | necessary for the normal response of Y. pestis | Adenosine triphosphate | 2 707 857|a|These findings show that LcrH is necessary for the normal response of Y. pestis to adenosine triphosphate and that LcrH contributes to Ca2+ respsonsiveness |
Samples of plague ontology data
| Subject . | Subtype . | ID . | Relation . | Object . | Obj-type . | ID . | Sentence . |
|---|---|---|---|---|---|---|---|
| Y. pestis | Bacteria | MeSH: D015010 | Sensitive | Gentamicin | Chemical | MeSH: D005839 | 29 312 891|a|Y. pestis is… |
| Plague | Disease | MeSH: D010930 | Causes die-off of colonies | Prairie dogs (Cynomys ludovicianus) | Species | 45 480 | 18 689 662|a|Plague, caused by… |
| YopM | Protein | GI: 1 946 646 191 | Necessary for virulence | Y. pestis | Bacteria | MeSH: D015010 | 2 401 564|t| YopM inhibits platelet… |
| Subject . | Subtype . | ID . | Relation . | Object . | Obj-type . | ID . | Sentence . |
|---|---|---|---|---|---|---|---|
| Y. pestis | Bacteria | MeSH: D015010 | Sensitive | Gentamicin | Chemical | MeSH: D005839 | 29 312 891|a|Y. pestis is… |
| Plague | Disease | MeSH: D010930 | Causes die-off of colonies | Prairie dogs (Cynomys ludovicianus) | Species | 45 480 | 18 689 662|a|Plague, caused by… |
| YopM | Protein | GI: 1 946 646 191 | Necessary for virulence | Y. pestis | Bacteria | MeSH: D015010 | 2 401 564|t| YopM inhibits platelet… |
Samples of plague ontology data
| Subject . | Subtype . | ID . | Relation . | Object . | Obj-type . | ID . | Sentence . |
|---|---|---|---|---|---|---|---|
| Y. pestis | Bacteria | MeSH: D015010 | Sensitive | Gentamicin | Chemical | MeSH: D005839 | 29 312 891|a|Y. pestis is… |
| Plague | Disease | MeSH: D010930 | Causes die-off of colonies | Prairie dogs (Cynomys ludovicianus) | Species | 45 480 | 18 689 662|a|Plague, caused by… |
| YopM | Protein | GI: 1 946 646 191 | Necessary for virulence | Y. pestis | Bacteria | MeSH: D015010 | 2 401 564|t| YopM inhibits platelet… |
| Subject . | Subtype . | ID . | Relation . | Object . | Obj-type . | ID . | Sentence . |
|---|---|---|---|---|---|---|---|
| Y. pestis | Bacteria | MeSH: D015010 | Sensitive | Gentamicin | Chemical | MeSH: D005839 | 29 312 891|a|Y. pestis is… |
| Plague | Disease | MeSH: D010930 | Causes die-off of colonies | Prairie dogs (Cynomys ludovicianus) | Species | 45 480 | 18 689 662|a|Plague, caused by… |
| YopM | Protein | GI: 1 946 646 191 | Necessary for virulence | Y. pestis | Bacteria | MeSH: D015010 | 2 401 564|t| YopM inhibits platelet… |
Graph storage and plague knowledge database construction
Based on the previous work, we have obtained the plague-related entities and their association relations. The above data are stored in Neo4j graph database. Subsequently, a plague knowledge graph called PlagueKG is constructed. The construction process comprises three parts:
Using scripts to classify entities and relations according to their types. Importing the classified data and other previously extracted knowledge, such as MESH numbers and sentence numbers, into an Excel file automatically;
Converting the file into .csv format and importing the nodes and relations into the graph database;
Starting the Neo4j graph database. The operations of addition, deletion, modification and checking can be conducted to the database.
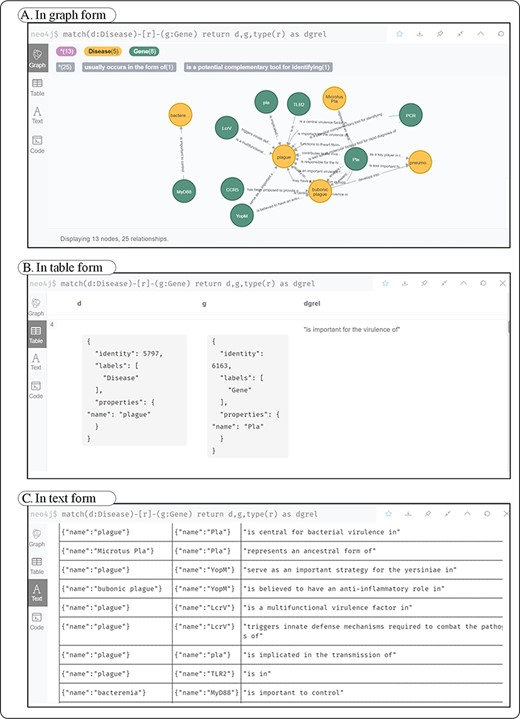
Neo4j provides three ways to display the query results through graphs, tables and texts. As shown in Figure 3, the correlations relations between plague and gene are revealed.
Three forms of disease–gene query results in Neo4j. (A) A kind of form in graph; (B) shows a kind of form in table; (C) a kind of form in text.
We built a plague knowledge database that contains a base layer, a data layer, a processing layer, a service layer and an application layer. The front end is built with Python + Django + web framework and back-end uses mysql + Neo4j graphical database to realize the management and visualization of the data.
Results
Plague knowledge graph
PlagueKG covers 9633 nodes of 33 types, 9446 relations of 292 types and 9583 triples connecting diseases, genes, proteins, symptoms, diagnosis, detection technology, equipment, geographical location and other entities. The knowledge graph presents plague-related knowledge in a visualized manner, thus allowing researchers to quickly understand this type of disease intuitively without spending much time and energy on reading literature. Accordingly, it provides a crucial reference point for researchers and may facilitate the development of a more effective vaccine.
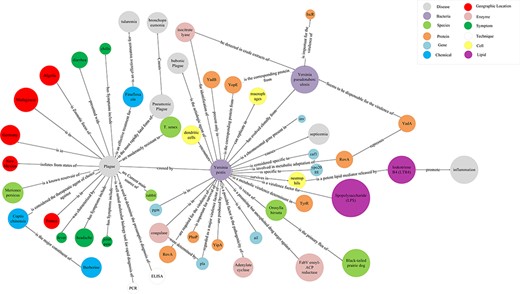
Because of the considerable number of entities and relations included in plague knowledge graph, detailed entities and relations information are difficult to present when the full picture is shown. Thus, the structure is illustrated through subgraph. Twelve types of entities and 55 relations are shown in Figure 4. Different colors represent different entity types, and the edges represent the relationships between entities.
Plague knowledge database
According to system framework requirements and research content, the database primarily designs four major functional modules: Plague Knowledge Graph Visualization Module, Plague Ontology Data Sheet Module, Text Auto-annotation Module and Manager Module. Among them, the Manager Module is subdivided into three submodules as follows: data upload, data management and Neo4j browser. The database can fully display the plague-related relations and provide convenient retrieval function for researchers, which is accessible on http://39.104.28.169:18095/. The home page of the system is shown in Figure 5.
Plague knowledge graph visualization module
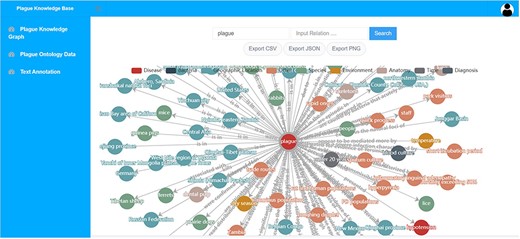
This module provides users with a visual presentation function for the plague knowledge graph. As depicted in Figure 6, users can access the data by directly querying entity or relation. The results can be exported in three formats: csv, json and png.
Plague data sheet module
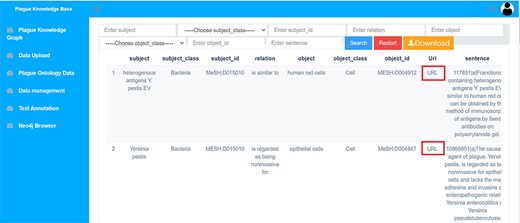
This module presents an information list of plague ontology base, containing entity categories, entity numbers, inter-entity relations as well as the corresponding sources of sentences. It is available for users to browse, search and download. Users are allowed to search and query in this module in accordance with their needs and filter table information. It also provides a drop-down list of entity types. Users are also allowed to query by type. Furthermore, the data can be exported in xlsx format.
Text annotation module
The major function of this module is to provide automatic annotation of biomedical concepts (i.e. chemical, species and proteins) in plague-related abstracts. The corresponding literature abstracts can be searched based on the PMID number, title or keyword. The search results suggest that different entity types in the text have been highlighted with different colors. As shown in Figure 7, the automatically annotated information includes the entity type and its unique NCBI_ID, thus providing a great convenience for researchers to access the information.
Manager module
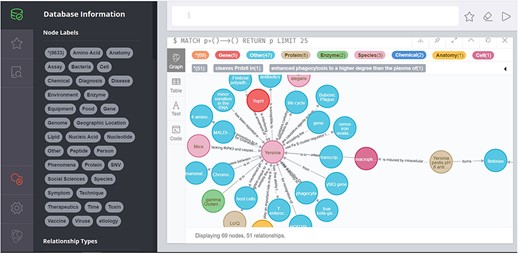
This module is developed for managers and biomedical experts. It covers three submodules: data upload, data management and Neo4j browser. It allows the manager to access the back-end to update and maintain the data on a regular basis. Data upload module provides the functions of uploading and exporting plague ontology data and plague text information in .xlsx format and .txt format, respectively, to keep the literature information updated with PubMed synchronously. The data management module provides a series of operations to add, delete, modify, query and update. As shown in Figure 8, the Neo4j Browser module provides accesses to the underlying data.
Plague knowledge graph visualization based on the Neo4j module.
Discussion
We use knowledge graph technology to logically relate the above fragmented knowledge in a graphical way, which intuitively and clearly presents a more comprehensive plague knowledge.
We are surprised to understand that in addition to the commonly known chemicals such as streptomycin, doxycycline and gentamycin, traditional Chinese herbs and therapies such as Coptis chinensis, Rheum officinale and moxibustion are effective in the prevention and treatment of plague. Accordingly, the treatment of plague by traditional Chinese medicine deserves further exploration. We have also learned that deltamethrin and primisulfuron are capable of eliminating fleas and cutting off the source of infection to control the spread of plague. Besides their role in the growth and metabolism of Y. pestis, enzymes can also be adopted to identify Y. pestis, isocitrate lyase is a good example. In addition, adenosine deaminase can distinguish between Y. pestis and pseudotuberculous microorganisms; Yersinia spores are found not only in traditional hosts, but also in our everyday foods such as raw meat, seafood and bottled water, thus revealing that our protection against plague should not be limited to traditional sources of infection.
Next, two case studies are conducted to demonstrate how PlagueKG can be adopted to facilitate biological discovery.
Case study
Case study 1: enzyme and bacteria
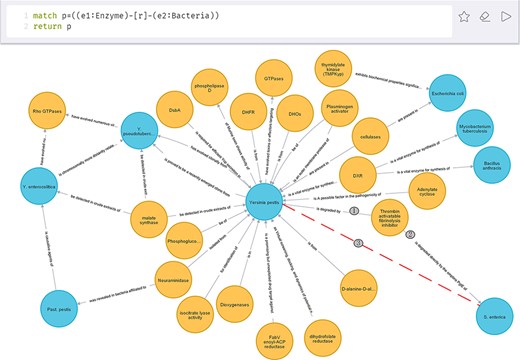
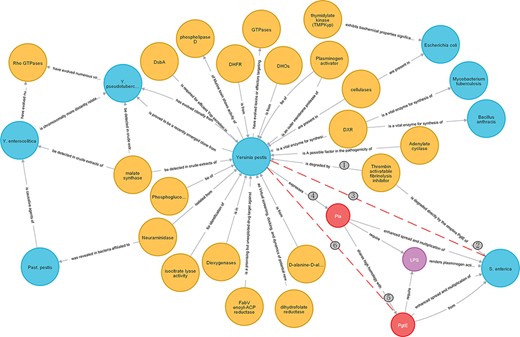
Enzyme takes on a critical significance in bacterial growth and metabolism. Researchers are very interested in it. Figure 9 shows the correlations between enzyme and bacteria after the execution of the cypher statement:’match p = ((e1:Enzyme)-[r]-(e2:Bacteria)) return p’. To be specific, yellow represents enzyme and blue represents bacteria.
As depicted in Figure 9, thrombin-activatable fibrinolysis inhibitor plays a crucial role in both Y. pestis and Salmonella enterica, thus suggesting that there may be some potential correlation between the two bacteria, as represented by the red dashed line, for example, whether the treatment of diseases caused by S. enterica can be used to treat Y. pestis deserves further exploration. Likewise, there may be a correlation between Y. pestis and Bacillus anthracis. Furthermore, isocitrate lyase can be employed for the identification of Y. pestis, such that it can be further recognized as a component of plague detection reagents.
Case study 2: expand relations
Researchers can also go deeper in accordance with Figure 9. As shown in Figure 10, red represents protein and purple represents lipid, when further expanding the relationship of S. enterica, and we can see that there is a correlation between Pla and PgtE. On that basis, it is speculated that there may be a potential correlation between PgtE and Y. pestis. For instance, PgtE may inhibit the infection and transmission of Y. pestis, which should be confirmed in depth.
To the best of our knowledge, this is the first plague knowledge database constructed by literature mining and knowledge graph techniques. PlagueKG places a greater focus showing the relationships between entities and allows data to be viewed and found visually and rapidly. Furthermore, it is considered that a more detailed and comprehensive data information can be more effectively presented in a list format, such that a list form is presented to show data in module of ‘Plague Ontology Data’, which allows researchers to index entities and inter-entity relationships, as well as by entity category, entity id and corresponding sentences. To ensure data traceability, users can click on the ‘URL’ button to jump to the PubMed link and view the corresponding article, as shown in Figure 11.
Conclusion and future work
In this paper, a total of 5388 literature related to plague are collected through literature mining. After named entity recognition and relation extraction, a plague knowledge graph is constructed that contains 33 entity types, 292 relation types, 9433 nodes and 9466 edges. Through the knowledge graph, we get a more intuitive and clear cognition of a series of plague-related biomedical concepts (i.e. chemicals, genes, species, proteins and diseases) and their correlations. The data are of great significance to the study of plague and provide reference and help for researchers. This study is likely to facilitate the vaccine development. Our plague knowledge database is capable of successfully managing and presenting considerable structured plague data, which is made available at http://39.104.28.169:18095/.
Since it takes a lot of time and energy to organize entities and relations data in a semi-automatic and semi-manual way, named entity recognition algorithms and relation extraction models will be proposed for plague in subsequent research. Moreover, intelligent question and answer algorithm and inference algorithm will be conducted in accordance with the constructed knowledge graph. Furthermore, the knowledge base will be enriched and updated regularly by adding novel plague-related data to provide researchers with a more practical and precise service.
Data availability
The data that support the findings of this study are available from http://39.104.28.169:18095/.
Authors’ contributions
J.G. designed and supervised the research work. J.L. conducted the biomedical text mining, wrote the manuscript and participated in the development of the knowledge database. B.F. conducted the development of knowledge database. Y.J. participated in some data collection and sorting.
Acknowledgements
We would like to express our sincere appreciation to the Professor Weiguang Zhou, an animal infectious disease expert from the Inner Mongolia Agricultural University and Director of Animal Infectious Diseases Branch of Chinese Society of Animal Husbandry and Veterinary Medicine and his team for their help and guidance in data collation and review.
Funding
The Inner Mongolia Science and Technology Major Special Projects (2019ZD016 and 2021ZD0005), Natural Science Foundation of Inner Mongolia Autonomous Region (2019MS03014) and Department of Education of Inner Mongolia Autonomous Region (S20210228Z).
Conflict of interest
The authors have no conflict of interest to declare.