-
PDF
- Split View
-
Views
-
Cite
Cite
Jinghang Gu, Emmanuele Chersoni, Xing Wang, Chu-Ren Huang, Longhua Qian, Guodong Zhou, LitCovid ensemble learning for COVID-19 multi-label classification, Database, Volume 2022, 2022, baac103, https://doi.org/10.1093/database/baac103
Close - Share Icon Share
Abstract
The Coronavirus Disease 2019 (COVID-19) pandemic has shifted the focus of research worldwide, and more than 10 000 new articles per month have concentrated on COVID-19–related topics. Considering this rapidly growing literature, the efficient and precise extraction of the main topics of COVID-19–relevant articles is of great importance. The manual curation of this information for biomedical literature is labor-intensive and time-consuming, and as such the procedure is insufficient and difficult to maintain. In response to these complications, the BioCreative VII community has proposed a challenging task, LitCovid Track, calling for a global effort to automatically extract semantic topics for COVID-19 literature. This article describes our work on the BioCreative VII LitCovid Track. We proposed the LitCovid Ensemble Learning (LCEL) method for the tasks and integrated multiple biomedical pretrained models to address the COVID-19 multi-label classification problem. Specifically, seven different transformer-based pretrained models were ensembled for the initialization and fine-tuning processes independently. To enhance the representation abilities of the deep neural models, diverse additional biomedical knowledge was utilized to facilitate the fruitfulness of the semantic expressions. Simple yet effective data augmentation was also leveraged to address the learning deficiency during the training phase. In addition, given the imbalanced label distribution of the challenging task, a novel asymmetric loss function was applied to the LCEL model, which explicitly adjusted the negative–positive importance by assigning different exponential decay factors and helped the model focus on the positive samples. After the training phase, an ensemble bagging strategy was adopted to merge the outputs from each model for final predictions. The experimental results show the effectiveness of our proposed approach, as LCEL obtains the state-of-the-art performance on the LitCovid dataset.
Database URL: https://github.com/JHnlp/LCEL
Introduction
The reality of the pandemic sweeping across the world and the challenges it has caused have rapidly accelerated the global pace of scientific publications (1, 2). Since more than 10 000 articles related to COVID-19 have been published monthly (3–5), the burden of manual curation and downstream interpretation has increased, making it difficult to access scientific analysis, pharmaceutical engineering and public usage.
In response to the rapid growth of COVID-19–related information, the LitCovid hub (3, 4), a new, curated literature database, has been developed to track up-to-date published research on COVID-19–related articles in PubMed. To facilitate the information retrieval, all curated publications in LitCovid are annotated by predefined semantic topics and updated daily. These elaborated semantic topics have been shown to be effective for various downstream applications such as citation analysis and knowledge graph generation (11). Currently, the annotation of biomedical semantic topics for COVID-19 literature is completed manually by human experts with specific domain knowledge (3, 4). However, the manual annotation of the fast-growing COVID-19 literature is labor-intensive and time-consuming. In order to assure accuracy, experts have to thoroughly examine the entirety of each biomedical article and assign it to a series of suitable predefined semantic topics. Moreover, the fact that biomedical literature often involves multiple topics rather than one single topic further aggravates the challenge of manual biocuration. Hence, the automatic curation and the interpretation of COVID-19 literature have become a problem of great importance.
Despite the preliminary efforts (6–10) providing feasible solutions in various knowledge extraction tasks for the biomedical domain, automated identification of COVID-19 semantic topics remains challenging. In addition, few identification tools for COVID-19 topics are freely available, and there are seldom successful examples of such applications in the real world.
In this regard, to tackle the automated topic identification for COVID-19, the BioCreative VII community stepped out and proposed the LitCovid challenge (11) in 2021. This task is regarded as a typical multi-label classification problem that calls for a global effort to provide practical benefits to worldwide biomedical curation. For this task, each participant was required to assign one or more semantic topics to each biomedical article. Each topic caters to different information needs of users and is effective for COVID-19–related information retrieval and downstream applications (3, 12). Particularly, seven specific COVID-19–related topics are proposed in the challenge, namely Treatment, Diagnosis, Prevention, Mechanism, Transmission, Case Report and Epidemic Forecasting. Table 1 presents the detailed descriptions of the semantic topics used in the LitCovid challenge.
Description of the semantic topics of the LitCovid challenge
| Topic . | Description . |
|---|---|
| Treatment | Treatment strategies, therapeutic procedures and vaccine development for COVID-19 |
| Diagnosis | COVID-19 assessment through symptoms, test results and radiological features for COVID-19 |
| Prevention | Prevention, control, mitigation and management strategies |
| Mechanism | Underlying cause(s) of COVID-19 infections and transmission and possible drug mechanism of action |
| Transmission | Characteristics and modes of COVID-19 transmissions |
| Case Report | Descriptions of specific patient cases related to COVID-19 |
| Epidemic Forecasting | Estimation on the trend of COVID-19 spread and related modeling approach |
| Topic . | Description . |
|---|---|
| Treatment | Treatment strategies, therapeutic procedures and vaccine development for COVID-19 |
| Diagnosis | COVID-19 assessment through symptoms, test results and radiological features for COVID-19 |
| Prevention | Prevention, control, mitigation and management strategies |
| Mechanism | Underlying cause(s) of COVID-19 infections and transmission and possible drug mechanism of action |
| Transmission | Characteristics and modes of COVID-19 transmissions |
| Case Report | Descriptions of specific patient cases related to COVID-19 |
| Epidemic Forecasting | Estimation on the trend of COVID-19 spread and related modeling approach |
Description of the semantic topics of the LitCovid challenge
| Topic . | Description . |
|---|---|
| Treatment | Treatment strategies, therapeutic procedures and vaccine development for COVID-19 |
| Diagnosis | COVID-19 assessment through symptoms, test results and radiological features for COVID-19 |
| Prevention | Prevention, control, mitigation and management strategies |
| Mechanism | Underlying cause(s) of COVID-19 infections and transmission and possible drug mechanism of action |
| Transmission | Characteristics and modes of COVID-19 transmissions |
| Case Report | Descriptions of specific patient cases related to COVID-19 |
| Epidemic Forecasting | Estimation on the trend of COVID-19 spread and related modeling approach |
| Topic . | Description . |
|---|---|
| Treatment | Treatment strategies, therapeutic procedures and vaccine development for COVID-19 |
| Diagnosis | COVID-19 assessment through symptoms, test results and radiological features for COVID-19 |
| Prevention | Prevention, control, mitigation and management strategies |
| Mechanism | Underlying cause(s) of COVID-19 infections and transmission and possible drug mechanism of action |
| Transmission | Characteristics and modes of COVID-19 transmissions |
| Case Report | Descriptions of specific patient cases related to COVID-19 |
| Epidemic Forecasting | Estimation on the trend of COVID-19 spread and related modeling approach |
In this article, building upon our previous research (13), we present the extension work of LitCovid Ensemble Learning (LCEL) for the challenge of BioCreative VII LitCovid Track. Specifically, we thoroughly explored seven different advanced pretrained models with heterogeneous architectures for ensemble learning, which guarantees the diversity and robustness of the deep neural networks. Moreover, to enhance the representation abilities of deep neural models, additional biomedical knowledge was proposed to facilitate the fruitfulness of the semantic expressions. Simple yet effective data augmentation was also exploited to address the learning deficiency during the training stage. In addition, to handle the imbalanced label distribution, a novel Asymmetric Loss (ASL) function (14) was introduced to the LCEL model, which explicitly adjusted the negative–positive importance by assigning different exponential decay factors. Benefiting from the usage of ASL, the proposed model was able to dynamically decouple the modulations of the positive and negative samples during the training phase and focused more on the positive samples while mitigating the contribution of negative ones.
The primary goal of this study was to develop a versatile machine learning approach with favorable robustness and generalizability to be easily applied to the COVID-19 domain and scaled up to other biomedical fields. The experimental results on the LitCovid dataset achieved state-of-the-art performance, demonstrating the effectiveness of our proposed method. The main contributions of this work are summarized as follows:
We propose a novel ensemble learning framework that can scale up effectively to the COVID-19 domain. Our study shows the superiority of the proposed method, which outperforms the current state-of-the-art systems.
We propose leveraging additional biomedical knowledge as well as data augmentation to enhance the semantic representation ability of the ensemble models. We argue that such kinds of semantic information can benefit the COVID-19 topic classification and supplement the development of relevant biomedical text mining technologies.
We introduce a novel loss function, ASL, which explicitly adjusts the importance of both positive and negative training samples. Due to the employment of ASL, the model can efficiently mitigate the imbalanced label distribution problem.
We make the related codes and materials of the proposed method publicly available to the research community. Our work is capable of offering new insights and building essential foundations for researchers in support of the ongoing fight against COVID-19.
Related work
In previous decades, biomedical topic identification was regarded as the multi-label classification problem, and a series of automated approaches (15–19) were developed to improve the time-consuming and labor-intensive curation process.
The National Library of Medicine (NLM) developed the most famous biomedical topic identification system, Medical Text Indexing (MTI) (15), which has been aiding the NLM human curators since 2002. The main purpose of MTI is to apply a rank-based approach to model the topic identification problem, where the top-ranked topics are recommended as true labels. Recently, with the comprehensive success of deep neural networks, deep learning–based approaches have brought remarkable breakthroughs in various biomedical topic identification tasks (16–19). FullMeSH (16) proposed a hybrid architecture integrating both deep neural networks and traditional machine learning methods to improve the topic identification performance. Specifically, it took advantage of Support Vector Machine, K-Nearest Neighbors algorithm and an attention-based convolution neural network to generate semantic evidence for the topic recommendation. Its attention mechanism exhibited remarkable potential by providing automatic feature representations without manual interference. AttentionMeSH (17) is another effective model based on attention mechanisms for biomedical topic identification. It utilized the architecture of a bidirectional recurrent neural network (RNN) with an attention mechanism to classify semantic topics for biomedical articles. Due to its deep representation capability, AttentionMeSH enabled the model to associate more textual evidence with candidate labels for better prediction results. MeSHProbeNet (18) and MeSHProbeNet-P (19) were two homogeneous deep learning methods that incorporated RNN and attention mechanisms simultaneously. By leveraging multiple semantic probes through an attention-based enhancement, MeSHProbeNet and MeSHProbeNet-P were able to acquire much deeper semantic insights into biomedical knowledge than the original documental contexts.
Despite preliminary efforts (15–19) that have provided feasible solutions and remarkable signs of progress over automatic topic identification, there is still an apparent gap between these automated methods and their applications to the COVID-19 domain. On the one hand, the above-described topic classification systems mainly concentrate on the topics of Medical Subject Headings (MeSH) (20, 21), which is a relatively large yet general set of biomedical concepts. Nevertheless, confronting the current pandemic crisis, there is a severe lack of such specialized topic collections targeting the evolving biomedical knowledge of COVID-19. On the other hand, lacking such a standard corpus for the COVID-19 domain drastically restricts the development of data mining techniques for identifying COVID-19–related semantic topics.
In light of these concerns, the BioCreative VII community proposed the challenging task of the LitCovid Track (11), which targets assigning multiple topic labels to COVID-19–relevant literature. This task is regarded as a typical multi-label classification problem that calls for a worldwide effort to provide practical benefits to COVID-19 biocuration. For this task, 19 teams participated and submitted a total of 80 valid predictions during the online competition. Pretraining methods dominated the challenging task and exhibited the best performance amid the online evaluation. Specifically, DUT914 (22) merged feature representations originating from different pretrained models to address the LitCovid multi-label classification problem. Likewise, DonutNLP (23) utilized a voting-based method integrating multiple pretrained models to enhance the representation ability for the final prediction. Our previous work, PolyU_CBSNLP (13), proposed to make full use of the homogeneous and heterogeneous structures of different pretrained models and achieved a promising performance during the online competition. Apart from the techniques of pretrained models, Bioformer (24) also exploited a large amount of external biomedical articles for further fine-tuning in the training phase, which helped the model achieve the best performance during the competition.
In view of multi-label classification, the characteristic is the inherent imbalanced positive–negative label distribution. This kind of task usually contains a relatively small portion of possible labels, implying that the number of positive samples per category will be, on average, much lower than that of negative ones. To address this issue, resampling-based methods are usually applied to balance the background positive–negative label distribution (25). However, such resampling methods are not always suitable for multi-label classification tasks, as the tasks contain multiple labels, while resampling cannot change the distribution of the specific label. A prominent solution for multi-label imbalance is to adopt the focal loss (26), which decays the loss as the label confidence increases. Focal loss helps the model focus on the hard samples while downweighting the easy ones, which demonstrates outstanding results in various object detection tasks. However, treating the positive and negative samples equally, as proposed by focal loss, is sometimes suboptimal, as it results in the accumulation of more loss gradients from negative samples while underemphasizing the importance of the rare positive ones. To this end, on the basis of focal loss (26), a novel ASL (14) emerges to operate differently on positive and negative samples. ASL enables deep neural models to dynamically downweight and hard threshold the easy negative samples, whereas the possibly mislabeled samples will be discarded. With the help of ASL, deep neural models achieved state-of-the-art performance on multiple popular image classification tasks (14). As there is a lack of such distinct research investigating the label imbalance for the COVID-19 domain, we propose introducing ASL to existing achievements to assist the research of COVID-19 topic identification.
Inspired by previous works, this article is devoted to the COVID-19 topic identification problem. We aim to provide a publicly available benchmark system with robust and flexible inherence for the COVID-19 domain, thus filling the important gap in previous research.
Dataset
In this section, we first present a brief introduction to the LitCovid corpus and then systematically depict the statistics of the corpus.
The LitCovid corpus developed by the BioCreative VII community originates from a large-scale curated literature hub, whose curated data are updated daily with the latest COVID-19–relevant articles; it is also publicly available for research purposes and industrial applications (3, 4). The BioCreative VII organizers collected more than 30 000 COVID-19–related articles from the literature hub (11), which were further split into three subsets of training, development and test datasets, respectively. During the competition phase, the organizers first released the training dataset as well as the development dataset in Comma-Separated Values format. Later, they released the test dataset following the same data schema except for the ground-truth labels, which were supposed to be predicted by the participants.
Since the LitCovid Track targeted the multi-label classification for COVID-19 semantic topics, seven specific topic labels were annotated in the corpus, namely Treatment, Diagnosis, Prevention, Mechanism, Transmission, Case Report and Epidemic Forecasting. Out of the semantic topics annotated for each article, the organizers also provided various kinds of metadata retrieved from PubMed, enhancing the fruitfulness of the dataset. More detailed information on the LitCovid corpus is shown in Tables 2 and 3.
The metadata statistics of the LitCovid corpus
| Metadata . | Train . | Development . | Test . |
|---|---|---|---|
| Title | 24 960 | 6239 | 2500 |
| Abstract | 24 900 | 6219 | 2485 |
| Journal name | 24 960 | 6239 | 2500 |
| Keywords | 18 968 | 4754 | 2056 |
| PMID | 24 960 | 6239 | 2500 |
| Authors | 24 859 | 6212 | 2499 |
| DOI | 24 406 | 6100 | 2474 |
| Publication type | 24 960 | 6239 | 2500 |
| Metadata . | Train . | Development . | Test . |
|---|---|---|---|
| Title | 24 960 | 6239 | 2500 |
| Abstract | 24 900 | 6219 | 2485 |
| Journal name | 24 960 | 6239 | 2500 |
| Keywords | 18 968 | 4754 | 2056 |
| PMID | 24 960 | 6239 | 2500 |
| Authors | 24 859 | 6212 | 2499 |
| DOI | 24 406 | 6100 | 2474 |
| Publication type | 24 960 | 6239 | 2500 |
The metadata statistics of the LitCovid corpus
| Metadata . | Train . | Development . | Test . |
|---|---|---|---|
| Title | 24 960 | 6239 | 2500 |
| Abstract | 24 900 | 6219 | 2485 |
| Journal name | 24 960 | 6239 | 2500 |
| Keywords | 18 968 | 4754 | 2056 |
| PMID | 24 960 | 6239 | 2500 |
| Authors | 24 859 | 6212 | 2499 |
| DOI | 24 406 | 6100 | 2474 |
| Publication type | 24 960 | 6239 | 2500 |
| Metadata . | Train . | Development . | Test . |
|---|---|---|---|
| Title | 24 960 | 6239 | 2500 |
| Abstract | 24 900 | 6219 | 2485 |
| Journal name | 24 960 | 6239 | 2500 |
| Keywords | 18 968 | 4754 | 2056 |
| PMID | 24 960 | 6239 | 2500 |
| Authors | 24 859 | 6212 | 2499 |
| DOI | 24 406 | 6100 | 2474 |
| Publication type | 24 960 | 6239 | 2500 |
The label distribution of the LitCovid corpus
| Label . | Train . | Development . | Test . |
|---|---|---|---|
| Treatment | 8718 | 2207 | 1035 |
| Diagnosis | 6193 | 1546 | 722 |
| Prevention | 11 102 | 2750 | 926 |
| Mechanism | 4439 | 1073 | 567 |
| Transmission | 1088 | 256 | 128 |
| Epidemic Forecasting | 645 | 192 | 41 |
| Case Report | 2063 | 482 | 197 |
| Total | 34 248 | 8506 | 3616 |
| Label . | Train . | Development . | Test . |
|---|---|---|---|
| Treatment | 8718 | 2207 | 1035 |
| Diagnosis | 6193 | 1546 | 722 |
| Prevention | 11 102 | 2750 | 926 |
| Mechanism | 4439 | 1073 | 567 |
| Transmission | 1088 | 256 | 128 |
| Epidemic Forecasting | 645 | 192 | 41 |
| Case Report | 2063 | 482 | 197 |
| Total | 34 248 | 8506 | 3616 |
The label distribution of the LitCovid corpus
| Label . | Train . | Development . | Test . |
|---|---|---|---|
| Treatment | 8718 | 2207 | 1035 |
| Diagnosis | 6193 | 1546 | 722 |
| Prevention | 11 102 | 2750 | 926 |
| Mechanism | 4439 | 1073 | 567 |
| Transmission | 1088 | 256 | 128 |
| Epidemic Forecasting | 645 | 192 | 41 |
| Case Report | 2063 | 482 | 197 |
| Total | 34 248 | 8506 | 3616 |
| Label . | Train . | Development . | Test . |
|---|---|---|---|
| Treatment | 8718 | 2207 | 1035 |
| Diagnosis | 6193 | 1546 | 722 |
| Prevention | 11 102 | 2750 | 926 |
| Mechanism | 4439 | 1073 | 567 |
| Transmission | 1088 | 256 | 128 |
| Epidemic Forecasting | 645 | 192 | 41 |
| Case Report | 2063 | 482 | 197 |
| Total | 34 248 | 8506 | 3616 |
Table 2 summarizes the basic statistical information of the LitCovid corpus. As shown in Table 2, there are 33 699 COVID-19–related biomedical articles in the corpus, with a training set of 24 960 articles, a development set of 6239 articles and a test set of 2500 articles. Most of the articles are filled with valid attributes of titles, abstracts, journal names, PubMed Identifiers (PMIDs), author names, Digital Object Identifiers (DOIs) as well as publication types. These abundant attributes guarantee the indispensable information which assures comprehensive coverage for research on COVID-19 topics and downstream applications. However, it is worth noting that despite the organizers trying their best to fill the metadata attributes, around 25% of keywords are still missing due to the incompleteness of the online information.
Table 3 depicts the label distribution of the LitCovid corpus. As shown in Table 3, it is observed that the frequency of different labels varies significantly. Among all topic labels, the labels of Prevention and Treatment dominate the entire corpus with relatively higher frequency, while the label of Epidemic Forecasting barely occurs, indicating an extremely imbalanced label distribution in the corpus, which makes the LitCovid challenge even harder, as most topic labels may never be observed in an article.
Methods
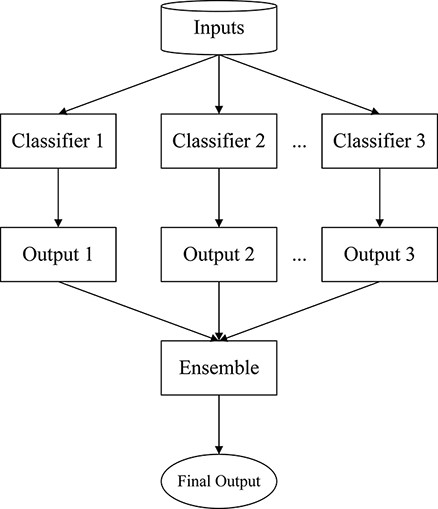
In this section, the LCEL paradigm is proposed for the COVID-19 multi-label classification problem. Figure 1 illustrates the architecture of the proposed method, which is a universal ensemble learning framework integrating multiple classifiers generated from different powerful pretrained models.
As known in the ensemble learning theory, every single model is taken as a weak learner or classifier due to its bias and variance in the feature representation (27). On this basis, the LCEL model is to train multiple weak classifiers separately through an ensemble manner and aggregate these weak classifiers into a stronger one to acquire better results. Specifically, we take advantage of multiple advanced pretrained models with different transformer-based structures for the initialization of LCEL (13). The hypothesis is that when weak classifiers aggregate appropriately, the system is able to efficiently narrow down the bias and variance of such weak learners to create a stronger learner, achieving a more accurate and robust performance.
In Figure 1, the overall framework of LCEL is shown. To begin, each classifier of the pretrained neural models is fine-tuned independently during the training process, and then all the outputs of these classifiers are merged through an ensemble bagging strategy to obtain the final topic prediction. Moreover, in order to improve the representation diversity and robustness of ensemble learning, the pretrained models with different architectural implementations are taken into consideration. Particularly, seven powerful pretrained transformers are elaborated in LCEL, i.e. PubMedBERT (28), CovidBERT (29), BioBERT-Large (30), BioBERT-Base (30), BioM-ELECTRA (31), BioELECTRA (32) and BioMed-RoBERTa (33). It is worth noticing that among these pretrained models, there are four variants of BERT (34), two variants of ELECTRA (35) and one edition of RoBERTa (36), respectively. We refer to the models with the same underlying architecture as homogeneous models; otherwise, the models are referred to as heterogeneous ones. These homogeneous and heterogeneous model groups ensure the effectiveness and stability of the proposed ensemble learning method.
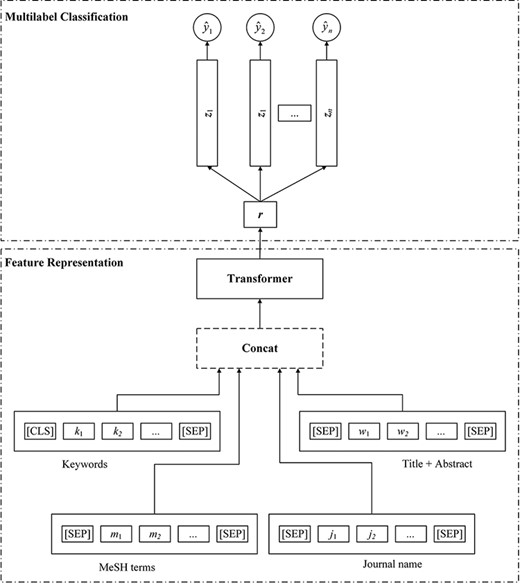
Figure 2 depicts the holistic structure of each pretrained classifier ensembled in LCEL. As shown in Figure 2, each classifier consists of two main modules, namely Feature Representation and Multi-label Classification. In this figure, the Feature Representation module takes the multiple textual components as the inputs and considers diverse semantic aspects for each input article. These textual inputs are then encoded by transformer-like encoders to generate further feature representations.
The structure of the transformer-based multi-label classifier.
After the Feature Representation stage, a linear classifier is adopted to take the extracted features from different semantic aspects to perform the final topic classification. For each candidate topic, the model is able to predict a probability score. In addition, to handle the imbalanced label distribution problem, a novel loss function, i.e., ASL, (14) is proposed in LCEL to dynamically adjust the learning weights between positive and negative instances during the training phase. More detailed information is described in the following subsections.
Feature representation
Since the titles and abstracts of biomedical literature convey rich contextual information that offers both explicit and implicit cues for determining topics, such contexts are regarded as the textual inputs for LCEL. However, despite the meaningful contexts of the biomedical literature, surface textual expressions are still less informative for semantic representation due to the deficiency of necessary knowledge comprehension.
Following our previous work (13), we argue that the additional biomedical knowledge, such as keywords, MeSH terms and journal names, is beneficial for the problem of COVID-19 topic identification. The main idea behind taking these kinds of additional biomedical knowledge is that they carry a large amount of manually refined semantic meanings that have been carefully reviewed by the authors and curators. Therefore, as shown at the bottom of Figure 2, before the training stage, the input sequence of each article needs to be constructed by concatenating all texts of keywords, MeSH terms, journal names, as well as titles and abstracts. Note that since the MeSH terms are not available in the official LitCovid corpus, we thus crawled these crucial materials as supplements from PubMed in terms of the corresponding identifier PMID of each target article.
where L is the length of the input sequence, |${\rm{\textbf{H}}} \in {\mathbb{R}^{{\rm{L}} \times {\rm{d}}}}$| is a L × d matrix concatenating all hidden states of the input words.
Nonetheless, although transformers-based methods have gained prominence in various NLP tasks (34, 37), their performance still highly relies on the quantity and diversity of the training data and usually suffers from the challenges of inadequate training samples. Moreover, deep neural models are also prone to overfit on small datasets due to their massive number of trainable parameters. In light of these concerns, when confronting the COVID-19 topic identification problem, a simple yet effective data augmentation approach is also introduced to LCEL for further performance improvement. Specifically, a large number of additional 96 804 COVID-19–relevant articles (including titles, abstracts, labels, etc.) are incorporated from the online hub (11) to enrich the representation capability of LCEL.
Multi-label classification
where |${\textrm{\textbf{W}}} \in {\mathbb{R}^{{\rm{m \times h}}}}$| is the linear transformation matrix, |${\textrm{\textbf{b}}} \in {\mathbb{R}^{\rm{m}}}$| is the bias and σ stands for the Sigmoid activation function. The value m equals the number of the target topics for the final classification, and each output can be interpreted as the confidence score of the corresponding topic recommendation.
Since the COVID-19 topic identification task is regarded as a multi-label classification problem, a high negative–positive label imbalance issue is inevitably encountered during the training phase. Therefore, how to tackle the extremely imbalanced label distribution is essential to the overall system performance. In previous decades, the conventional loss function of binary cross-entropy (BCE) has dominated various challenging NLP tasks and similarly exhibited remarkable success in numerous biomedical fields (16–19). However, the aggressive essence of BCE treats all candidate labels equally without distinguishing the different importance between positive and negative samples. This disadvantage usually leads to suboptimal performance (14, 26), as it results in the accumulation of more loss gradients from negative samples while downweighting the importance of the rare positive ones.
where p = σ(z) is the confidence probability predicted by the network, pm is the shifted probability and γ+ and γ- are the asymmetric focusing weights decaying the contribution of both positive and negative samples, respectively. The probability margin m ≥ 0 is a tunable hyper-parameter that controls the acceptance of the negative predictions.
It is observed that ASL decouples the focusing levels of positive and negative samples through different decay rates γ+ and γ-. When dealing with multi-label training, higher values of decay rates are able to sufficiently downweight the contribution from easy samples (14). Specifically, since the number of negative samples is much larger than the positive ones in the corpus, by setting γ− > 0 in Equation (6), the contribution of easy negatives (with low probability, p ≪ 0.5) to the accumulated loss can be significantly downweighted, enabling the model to focus more on the harder samples during training. Moreover, the additional asymmetric mechanism of probability shifting also performs hard thresholding for the easy negative samples, i.e. it will fully discard the negative samples via a flexible probability margin m when their predicted confidence is very low. In short, through asymmetric focusing and probability shifting, ASL is able to obtain better control over the contributions of positive and negative samples to the loss function and help the model to learn meaningful features from positive samples despite their less frequent occurrences.
It is worth noting that when γ+ = γ- = pm = 0, ASL degenerates to the classic BCE loss. Since we are more interested in highlighting the contribution of positive samples, we set γ− > γ+ for experimentation. It can be convenient to set γ+ = 0 so that the positive samples will incur the same cross-entropy loss; meanwhile, the model only needs to control the level of the asymmetrically negative part via the hyper-parameter γ−. In other words, the model is able to focus on learning features from difficult samples while de-emphasizing the features from the easy ones.
Benefiting from the effectiveness of ASL, the issue of imbalanced label distribution is lessened and the entire framework of LCEL is trained in an end-to-end fashion by a gradient-based optimization algorithm that minimizes the total loss of |${\mathcal{L}_{{\rm{all}}}}$|.
Results
In this section, we first introduce the evaluation metrics and the experimental settings for the LitCovid multi-label classification problem; we then systematically evaluate the performance of our approach and compare it with the relevant state-of-the-art systems. Finally, the error analysis is carried out at the end of this section.
Evaluation metrics
Amid the LitCovid online competition, all submissions were evaluated from the label-based perspective and the instance-based perspective, both of which are widely utilized for multi-label classification. Specifically, nine different measures at three different levels are applied, i.e. example-based precision (EBP), example-based recall (EBR), example-based F1 (EBF), macro-precision (MaP), macro-recall (MaR), macro-F1 (MaF), micro-precision (MiP), micro-recall (MiR) and micro-F1 (MiF).
Let K denote the total number of all topic labels, and N denote the number of the input instances (i.e. biomedical articles). yi and |${\hat {\textrm{\textbf{y}}}_{\rm{i}}} \in {\left\{ {0,1} \right\}^{\rm{K}}}$| are the true and predicted labels for instance i, respectively. The foregoing evaluation metrics are further defined as follows:
EBF:
where
Note that EBP and EBR are calculated by summing EBPi and EBRi over all instances, respectively.
MaF
The MaP and MaR are obtained by computing the precision and recall for each label separately and then averaging them over all labels, as follows:
where
MiF
where
Experimental settings
In our experiments, all texts of articles and additional biomedical knowledge are converted into lower cases before being fed into the downstream deep neural networks. In case some article texts might exceed the length limitations of pretrained models, the overlong texts of biomedical articles are truncated to substitute the original ones. In our experiments, the maximum length of input texts is fixed to 512, and the training batch size is set to 50. As multiple state-of-the-art pretrained models are explored in LCEL, all parameters follow their default settings during the model initialization. In the training phase, the AdamW optimizer (38) is adopted to minimize the training loss, and the learning rates are kept identically for all models with the value of 2e-5.
Regarding the loss function of ASL, since we are more interested in emphasizing the contributions of positive samples, we empirically set γ− > γ+ to explicitly minimize the weights of negative samples. In particular, γ− and γ+ are separately assigned to 1 and 0. To simplify the experimentations, the hyper-parameter of the probability margin m follows the default settings as mentioned in the study by Ben-Baruch et al. (14), which equals 0.05. In terms of data augmentation, an additional 96 804 COVID-19 relevant articles are collected from the online hub (11) on 25 January 2022, excluding the overlaps with the original LitCovid dataset.
The total number of fine-tuning steps for each pretrained model is set to 16 000, and each checkpoint per 500 training steps is reserved for further evaluation and integration. The best-performed checkpoints of each pretrained model will be ensembled for the final topic prediction.
System performance on the development dataset
Our experimental results on the LitCovid development dataset are presented in the following order:
Evaluation with different biomedical knowledge on the LitCovid development dataset.
Evaluation with ASL and the performance comparison with BCE loss.
Evaluation with data augmentation and the overall comparison with different training policies.
System performance with different knowledge features
Following the previous work (13), three distinctive kinds of additional biomedical knowledge are proposed to enhance the feature representations for LCEL. To investigate the importance of the proposed biomedical knowledge, a detailed feature combination study is performed on the LitCovid development dataset, trying to reveal their different influences. Since seven advanced pretrained models are proposed for the ensemble learning of LCEL, the naive yet effective pretrained model of PubMedBERT (28) is selected to simplify the experimental comparison. The feature combination study follows the same training scenario described in the study by Gu et al. (13), i.e. it only utilizes the default LitCovid training dataset as well as BCE loss for fine-tuning. Table 4 depicts the details of the knowledge combination experiments, in which the best scores are highlighted in boldface. It is worth mentioning that all experiments rely on the fundamental texts (i.e. titles and abstracts) of the input articles, which are available for all kinds of trials.
The knowledge combination experiments on the development dataset
| Additional biomedical knowledge . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Text + Keywords + MeSH + Journal name | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| Text + Keywords + MeSH | 85.73 | 86.34 | 86.02 | 90.19 | 90.12 | 90.16 | 92.37 | 92.58 | 92.47 |
| Text + Keywords | 85.80 | 85.79 | 85.76 | 90.34 | 89.69 | 90.01 | 92.37 | 92.26 | 92.31 |
| Text | 85.14 | 84.17 | 84.60 | 90.10 | 89.22 | 89.66 | 92.09 | 91.83 | 91.96 |
| Additional biomedical knowledge . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Text + Keywords + MeSH + Journal name | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| Text + Keywords + MeSH | 85.73 | 86.34 | 86.02 | 90.19 | 90.12 | 90.16 | 92.37 | 92.58 | 92.47 |
| Text + Keywords | 85.80 | 85.79 | 85.76 | 90.34 | 89.69 | 90.01 | 92.37 | 92.26 | 92.31 |
| Text | 85.14 | 84.17 | 84.60 | 90.10 | 89.22 | 89.66 | 92.09 | 91.83 | 91.96 |
The knowledge combination experiments on the development dataset
| Additional biomedical knowledge . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Text + Keywords + MeSH + Journal name | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| Text + Keywords + MeSH | 85.73 | 86.34 | 86.02 | 90.19 | 90.12 | 90.16 | 92.37 | 92.58 | 92.47 |
| Text + Keywords | 85.80 | 85.79 | 85.76 | 90.34 | 89.69 | 90.01 | 92.37 | 92.26 | 92.31 |
| Text | 85.14 | 84.17 | 84.60 | 90.10 | 89.22 | 89.66 | 92.09 | 91.83 | 91.96 |
| Additional biomedical knowledge . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Text + Keywords + MeSH + Journal name | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| Text + Keywords + MeSH | 85.73 | 86.34 | 86.02 | 90.19 | 90.12 | 90.16 | 92.37 | 92.58 | 92.47 |
| Text + Keywords | 85.80 | 85.79 | 85.76 | 90.34 | 89.69 | 90.01 | 92.37 | 92.26 | 92.31 |
| Text | 85.14 | 84.17 | 84.60 | 90.10 | 89.22 | 89.66 | 92.09 | 91.83 | 91.96 |
It is observed from Table 4 that, by merely using the contextual information of titles and abstracts, PubMedBERT (28) is able to achieve an MaF as high as 84.60%, an MiF as high as 89.66% and an EBF as high as 91.96%, respectively. This indicates that the contextual information of titles and abstracts inherently contains crucial clues for COVID-19 multi-label classification, and the pretrained model can effectively capture and represent such useful information. When successively combing the contexts with the biomedical knowledge of keywords and MeSH terms, the performance is further improved and reaches an MaF of 86.02%, an MiF of 90.16% and an EBF of 92.47%, respectively. This suggests that these distinctive kinds of biomedical knowledge can significantly provide supplementary semantic information for COVID-19–relevant topics, which have been manually refined and interpreted by authors and curators. Interestingly, with further combing of the knowledge of journal names, the model is able to obtain the highest EBF of 92.61%, while losing slight performance in MaF and MiF. This indicates that the journal name does bring certain background knowledge to the pretrained model; however, such biomedical knowledge is too general to help the model improve overall. Although there are some slight losses in MaF and MiF, the knowledge of journal names still helps the model perform consistently better than the one that exclusively uses the contexts of titles and abstracts. In this regard, we take all biomedical knowledge of keywords, MeSH terms and journal names as the default features for all pretrained models during our experimentation.
System performance with ASL
As ASL is proposed for better control of the contributions from both positive and negative samples, a fair comparison between ASL and BCE is conducted to better understand the difference and to demonstrate the influence of the asymmetric focusing mechanism. Similar to the aforementioned feature combination, the single model of PubMedBERT (28) on the LitCovid development dataset is evaluated to simplify the experimental comparison. Table 5 reports the detailed performance of each label and their averaged results in micro, macro and example-based measures, respectively.
The comparison of different loss functions
| Loss . | Labels . | P (%) . | R (%) . | F1 (%) . | Count . |
|---|---|---|---|---|---|
| BCE | Treatment | 89.74 | 90.76 | 90.25 | 2207 |
| Diagnosis | 85.27 | 90.62 | 87.86 | 1546 | |
| Prevention | 94.06 | 95.09 | 94.58 | 2750 | |
| Mechanism | 88.86 | 86.21 | 87.51 | 1073 | |
| Transmission | 70.22 | 74.61 | 72.35 | 256 | |
| Epidemic Forecasting | 74.16 | 80.73 | 77.31 | 192 | |
| Case Report | 90.67 | 92.74 | 91.69 | 482 | |
| macro avg | 84.71 | 87.25 | 85.93 | 8506 | |
| micro avg | 89.24 | 90.96 | 90.09 | 8506 | |
| example avg | 92.02 | 93.20 | 92.61 | 6239 | |
| ASL | Treatment | 89.08 | 91.30 | 90.18 | 2207 |
| Diagnosis | 87.94 | 90.56 | 89.23 | 1546 | |
| Prevention | 93.92 | 96.11 | 95.00 | 2750 | |
| Mechanism | 88.39 | 87.23 | 87.80 | 1073 | |
| Transmission | 70.00 | 68.36 | 69.17 | 256 | |
| Epidemic Forecasting | 76.88 | 79.69 | 78.26 | 192 | |
| Case Report | 92.23 | 91.08 | 91.65 | 482 | |
| macro avg | 85.49 | 86.33 | 85.90 | 8506 | |
| micro avg | 89.70 | 91.24 | 90.47 | 8506 | |
| example avg | 92.43 | 93.43 | 92.93 | 6239 |
| Loss . | Labels . | P (%) . | R (%) . | F1 (%) . | Count . |
|---|---|---|---|---|---|
| BCE | Treatment | 89.74 | 90.76 | 90.25 | 2207 |
| Diagnosis | 85.27 | 90.62 | 87.86 | 1546 | |
| Prevention | 94.06 | 95.09 | 94.58 | 2750 | |
| Mechanism | 88.86 | 86.21 | 87.51 | 1073 | |
| Transmission | 70.22 | 74.61 | 72.35 | 256 | |
| Epidemic Forecasting | 74.16 | 80.73 | 77.31 | 192 | |
| Case Report | 90.67 | 92.74 | 91.69 | 482 | |
| macro avg | 84.71 | 87.25 | 85.93 | 8506 | |
| micro avg | 89.24 | 90.96 | 90.09 | 8506 | |
| example avg | 92.02 | 93.20 | 92.61 | 6239 | |
| ASL | Treatment | 89.08 | 91.30 | 90.18 | 2207 |
| Diagnosis | 87.94 | 90.56 | 89.23 | 1546 | |
| Prevention | 93.92 | 96.11 | 95.00 | 2750 | |
| Mechanism | 88.39 | 87.23 | 87.80 | 1073 | |
| Transmission | 70.00 | 68.36 | 69.17 | 256 | |
| Epidemic Forecasting | 76.88 | 79.69 | 78.26 | 192 | |
| Case Report | 92.23 | 91.08 | 91.65 | 482 | |
| macro avg | 85.49 | 86.33 | 85.90 | 8506 | |
| micro avg | 89.70 | 91.24 | 90.47 | 8506 | |
| example avg | 92.43 | 93.43 | 92.93 | 6239 |
The comparison of different loss functions
| Loss . | Labels . | P (%) . | R (%) . | F1 (%) . | Count . |
|---|---|---|---|---|---|
| BCE | Treatment | 89.74 | 90.76 | 90.25 | 2207 |
| Diagnosis | 85.27 | 90.62 | 87.86 | 1546 | |
| Prevention | 94.06 | 95.09 | 94.58 | 2750 | |
| Mechanism | 88.86 | 86.21 | 87.51 | 1073 | |
| Transmission | 70.22 | 74.61 | 72.35 | 256 | |
| Epidemic Forecasting | 74.16 | 80.73 | 77.31 | 192 | |
| Case Report | 90.67 | 92.74 | 91.69 | 482 | |
| macro avg | 84.71 | 87.25 | 85.93 | 8506 | |
| micro avg | 89.24 | 90.96 | 90.09 | 8506 | |
| example avg | 92.02 | 93.20 | 92.61 | 6239 | |
| ASL | Treatment | 89.08 | 91.30 | 90.18 | 2207 |
| Diagnosis | 87.94 | 90.56 | 89.23 | 1546 | |
| Prevention | 93.92 | 96.11 | 95.00 | 2750 | |
| Mechanism | 88.39 | 87.23 | 87.80 | 1073 | |
| Transmission | 70.00 | 68.36 | 69.17 | 256 | |
| Epidemic Forecasting | 76.88 | 79.69 | 78.26 | 192 | |
| Case Report | 92.23 | 91.08 | 91.65 | 482 | |
| macro avg | 85.49 | 86.33 | 85.90 | 8506 | |
| micro avg | 89.70 | 91.24 | 90.47 | 8506 | |
| example avg | 92.43 | 93.43 | 92.93 | 6239 |
| Loss . | Labels . | P (%) . | R (%) . | F1 (%) . | Count . |
|---|---|---|---|---|---|
| BCE | Treatment | 89.74 | 90.76 | 90.25 | 2207 |
| Diagnosis | 85.27 | 90.62 | 87.86 | 1546 | |
| Prevention | 94.06 | 95.09 | 94.58 | 2750 | |
| Mechanism | 88.86 | 86.21 | 87.51 | 1073 | |
| Transmission | 70.22 | 74.61 | 72.35 | 256 | |
| Epidemic Forecasting | 74.16 | 80.73 | 77.31 | 192 | |
| Case Report | 90.67 | 92.74 | 91.69 | 482 | |
| macro avg | 84.71 | 87.25 | 85.93 | 8506 | |
| micro avg | 89.24 | 90.96 | 90.09 | 8506 | |
| example avg | 92.02 | 93.20 | 92.61 | 6239 | |
| ASL | Treatment | 89.08 | 91.30 | 90.18 | 2207 |
| Diagnosis | 87.94 | 90.56 | 89.23 | 1546 | |
| Prevention | 93.92 | 96.11 | 95.00 | 2750 | |
| Mechanism | 88.39 | 87.23 | 87.80 | 1073 | |
| Transmission | 70.00 | 68.36 | 69.17 | 256 | |
| Epidemic Forecasting | 76.88 | 79.69 | 78.26 | 192 | |
| Case Report | 92.23 | 91.08 | 91.65 | 482 | |
| macro avg | 85.49 | 86.33 | 85.90 | 8506 | |
| micro avg | 89.70 | 91.24 | 90.47 | 8506 | |
| example avg | 92.43 | 93.43 | 92.93 | 6239 |
In Table 5, it is noticeable that when adopting the conventional BCE loss, the best performance of PubMedBERT (28), on average, achieves an MaF of 85.93%, an MiF of 90.09% and an EBF of 92.61%. This indicates that the conventional BCE loss is universal and robust enough to handle the multi-label classification problem, due to its equivalent weight estimation over different label distributions. In contrast, when using ASL, the performance surpasses the model with BCE loss by improvements of 0.38 units in MiF and 0.32 units in EBF. This suggests that ASL enables the model to decouple the modulations of positive and negative samples, and the asymmetric focusing mechanism helps the model to understand positive samples better. However, when applying ASL, the model suffers from a slight decline in MaF with 0.03 units lower than the one with BCE. This implies that, although the asymmetric focusing mechanism concentrates more on the low-distributional positive samples, the aggressive adjustments of the weight manipulation might harm the importance of certain labels.
Specifically, by adopting BCE, the prediction of Prevention achieves the highest F1 score of 94.58%. The performance scores of Treatment, Diagnosis, Mechanism and Case Report are relatively close. Compared to the above-mentioned labels, predictions of Epidemic Forecasting and Transmission perform the worst. This is likely due to the label imbalance described in Section Dataset, which implies that with fewer class examples, the model faces more difficulties during prediction. Benefiting from the asymmetric focusing mechanism, ASL successfully helps the model pay more attention to the labels of Diagnosis, Prevention, Mechanism and Epidemic Forecasting, gaining better performance in the multi-label classification.
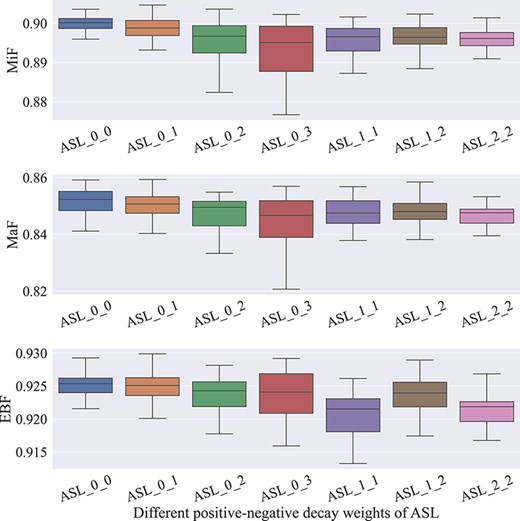
To understand the impacts of the exponential decay factors in the asymmetric focusing mechanism, we combine γ+ and γ− with different values to verify their influences. Note that for each pair of decay factors, we fine-tune the pretrained model and save multiple checkpoints for evaluation as described in the Section Experimental Settings. Figure 3 illustrates the boxplots of the standardized five-number summary for all checkpoints of pretrained models when using different settings of γ+ and γ−. Specifically, the minimum, the maximum, the sample median and the first and third quartiles are depicted in Figure 3. During the comparison, we set γ+ with the range from 0 to 2 and γ− with the range from 0 to 3, respectively. When tuning γ+ and γ-, all other parameters of pretrained models remain the same.
It is observed from Figure 3 that in all experimental trials, the best-performing checkpoint is gained when adopting the combination of the decay factors with γ+ = 0 and γ- = 1 (i.e. ASL_0_1). This can be explained by the advantages of the asymmetric focusing mechanism, which focuses more on the positive samples while attenuating the importance of negative ones. Interestingly, when simply applying γ+ = 0 and γ- = 0 (i.e. ASL_0_0), the best checkpoint of the model also exhibits comparable performance, which implies that the equal treatment of both positive and negative samples is as effective as the inherent assumption of BCE.
However, when γ+ is fixed to 0 and γ- is above 1, the pretrained models perform much worse. This is likely due to excessive downweighting of γ-, which may lead to too much disregard for negative samples, losing the necessary semantic information for the models. It is also noticeable that as the asymmetric decay weight γ- becomes higher, the prediction variance of the model also increases. This may further support the importance of keeping modest magnitudes of the contributions from both positive and negative samples. Moreover, allowing γ+ > 0, all the pretrained models achieve suboptimal performance, demonstrating that too much attenuation on the positive samples cannot provide more meaningful clues for further improvement.
System performance with data augmentation
Likewise, data augmentation is another approach proposed in LCEL that aims at benefiting the representation capabilities of pretrained models. To investigate the importance of corresponding contributions of data augmentation, we experiment with different training policies and compare their results, as seen in Table 6. One of the key claims is that data augmentation is able to provide meaningful background information that is crucial for COVID-19 multi-label classification. To verify the assumption, Table 6 exhibits the details of the experiments with different training policies.
The overall comparison of data augmentation with different loss functions
| Policy . | Model . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|---|
| Train_Def + BCE | PubMedBERT | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| CovidBERT | 84.65 | 85.23 | 84.86 | 88.51 | 90.45 | 89.47 | 91.30 | 92.68 | 91.98 | |
| BioBERT-Large | 85.20 | 78.01 | 80.71 | 88.39 | 87.54 | 87.96 | 90.34 | 90.23 | 90.28 | |
| BioBERT-Base | 84.12 | 82.09 | 82.71 | 88.56 | 88.28 | 88.42 | 91.02 | 90.87 | 90.94 | |
| BioM-ELECTRA | 86.45 | 83.76 | 85.02 | 89.68 | 90.04 | 89.86 | 92.32 | 92.60 | 92.46 | |
| BioELECTRA | 83.34 | 85.77 | 84.48 | 88.59 | 90.81 | 89.68 | 91.68 | 93.09 | 92.38 | |
| BioMed_RoBERTa | 85.18 | 85.48 | 85.32 | 89.52 | 90.87 | 90.19 | 92.19 | 93.12 | 92.65 | |
| Train_Def + ASL | PubMedBERT | 85.49 | 86.33 | 85.90 | 89.70 | 91.24 | 90.47 | 92.43 | 93.43 | 92.93 |
| CovidBERT | 84.57 | 85.17 | 84.83 | 88.95 | 90.50 | 89.72 | 91.59 | 92.81 | 92.20 | |
| BioBERT-Large | 82.63 | 81.58 | 82.06 | 88.11 | 87.64 | 87.88 | 90.54 | 90.61 | 90.57 | |
| BioBERT-Base | 85.87 | 86.32 | 86.07 | 90.27 | 90.62 | 90.44 | 92.65 | 93.04 | 92.84 | |
| BioM-ELECTRA | 83.73 | 87.06 | 85.34 | 88.92 | 91.89 | 90.38 | 91.95 | 93.92 | 92.92 | |
| BioELECTRA | 83.67 | 87.43 | 85.47 | 88.61 | 91.49 | 90.03 | 91.70 | 93.50 | 92.59 | |
| BioMed_RoBERTa | 84.32 | 87.27 | 85.75 | 88.75 | 91.68 | 90.19 | 91.76 | 93.69 | 92.71 | |
| Train _Aug + BCE | PubMedBERT | 86.47 | 86.62 | 86.51 | 90.38 | 91.58 | 90.98 | 92.88 | 93.74 | 93.31 |
| CovidBERT | 85.26 | 86.62 | 85.62 | 89.74 | 91.36 | 90.54 | 92.45 | 93.48 | 92.96 | |
| BioBERT-Large | 85.88 | 81.02 | 83.33 | 90.54 | 87.56 | 89.03 | 92.13 | 90.64 | 91.38 | |
| BioBERT-Base | 88.16 | 85.14 | 86.45 | 90.98 | 91.46 | 91.22 | 93.04 | 93.49 | 93.26 | |
| BioM-ELECTRA | 87.94 | 85.39 | 86.52 | 90.70 | 91.24 | 90.97 | 92.88 | 93.43 | 93.15 | |
| BioELECTRA | 84.73 | 89.24 | 86.83 | 89.70 | 92.05 | 90.86 | 92.21 | 94.04 | 93.12 | |
| BioMed_RoBERTa | 89.10 | 83.62 | 86.11 | 91.76 | 90.21 | 90.98 | 93.40 | 92.54 | 92.97 | |
| Train _Aug + ASL | PubMedBERT | 84.86 | 87.93 | 86.36 | 89.75 | 92.45 | 91.08 | 92.54 | 94.34 | 93.43 |
| CovidBERT | 84.98 | 87.10 | 86.01 | 89.68 | 91.74 | 90.70 | 92.41 | 93.79 | 93.09 | |
| BioBERT-Large | 82.59 | 86.73 | 84.52 | 87.27 | 91.24 | 89.21 | 90.58 | 93.28 | 91.91 | |
| BioBERT-Base | 85.02 | 88.11 | 86.51 | 89.69 | 92.24 | 90.95 | 92.50 | 94.13 | 93.31 | |
| BioM-ELECTRA | 84.90 | 88.56 | 86.55 | 89.80 | 92.36 | 91.06 | 92.62 | 94.16 | 93.38 | |
| BioELECTRA | 84.92 | 87.88 | 86.36 | 89.22 | 92.65 | 90.90 | 92.12 | 94.45 | 93.27 | |
| BioMed_RoBERTa | 84.36 | 89.25 | 86.67 | 87.90 | 93.82 | 90.76 | 91.52 | 95.31 | 93.38 | |
| LCEL¬ (Train _Aug + ASL) | Ensembled | 86.37 | 88.45 | 87.40 | 90.53 | 93.00 | 91.75 | 93.08 | 94.76 | 93.91 |
| Policy . | Model . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|---|
| Train_Def + BCE | PubMedBERT | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| CovidBERT | 84.65 | 85.23 | 84.86 | 88.51 | 90.45 | 89.47 | 91.30 | 92.68 | 91.98 | |
| BioBERT-Large | 85.20 | 78.01 | 80.71 | 88.39 | 87.54 | 87.96 | 90.34 | 90.23 | 90.28 | |
| BioBERT-Base | 84.12 | 82.09 | 82.71 | 88.56 | 88.28 | 88.42 | 91.02 | 90.87 | 90.94 | |
| BioM-ELECTRA | 86.45 | 83.76 | 85.02 | 89.68 | 90.04 | 89.86 | 92.32 | 92.60 | 92.46 | |
| BioELECTRA | 83.34 | 85.77 | 84.48 | 88.59 | 90.81 | 89.68 | 91.68 | 93.09 | 92.38 | |
| BioMed_RoBERTa | 85.18 | 85.48 | 85.32 | 89.52 | 90.87 | 90.19 | 92.19 | 93.12 | 92.65 | |
| Train_Def + ASL | PubMedBERT | 85.49 | 86.33 | 85.90 | 89.70 | 91.24 | 90.47 | 92.43 | 93.43 | 92.93 |
| CovidBERT | 84.57 | 85.17 | 84.83 | 88.95 | 90.50 | 89.72 | 91.59 | 92.81 | 92.20 | |
| BioBERT-Large | 82.63 | 81.58 | 82.06 | 88.11 | 87.64 | 87.88 | 90.54 | 90.61 | 90.57 | |
| BioBERT-Base | 85.87 | 86.32 | 86.07 | 90.27 | 90.62 | 90.44 | 92.65 | 93.04 | 92.84 | |
| BioM-ELECTRA | 83.73 | 87.06 | 85.34 | 88.92 | 91.89 | 90.38 | 91.95 | 93.92 | 92.92 | |
| BioELECTRA | 83.67 | 87.43 | 85.47 | 88.61 | 91.49 | 90.03 | 91.70 | 93.50 | 92.59 | |
| BioMed_RoBERTa | 84.32 | 87.27 | 85.75 | 88.75 | 91.68 | 90.19 | 91.76 | 93.69 | 92.71 | |
| Train _Aug + BCE | PubMedBERT | 86.47 | 86.62 | 86.51 | 90.38 | 91.58 | 90.98 | 92.88 | 93.74 | 93.31 |
| CovidBERT | 85.26 | 86.62 | 85.62 | 89.74 | 91.36 | 90.54 | 92.45 | 93.48 | 92.96 | |
| BioBERT-Large | 85.88 | 81.02 | 83.33 | 90.54 | 87.56 | 89.03 | 92.13 | 90.64 | 91.38 | |
| BioBERT-Base | 88.16 | 85.14 | 86.45 | 90.98 | 91.46 | 91.22 | 93.04 | 93.49 | 93.26 | |
| BioM-ELECTRA | 87.94 | 85.39 | 86.52 | 90.70 | 91.24 | 90.97 | 92.88 | 93.43 | 93.15 | |
| BioELECTRA | 84.73 | 89.24 | 86.83 | 89.70 | 92.05 | 90.86 | 92.21 | 94.04 | 93.12 | |
| BioMed_RoBERTa | 89.10 | 83.62 | 86.11 | 91.76 | 90.21 | 90.98 | 93.40 | 92.54 | 92.97 | |
| Train _Aug + ASL | PubMedBERT | 84.86 | 87.93 | 86.36 | 89.75 | 92.45 | 91.08 | 92.54 | 94.34 | 93.43 |
| CovidBERT | 84.98 | 87.10 | 86.01 | 89.68 | 91.74 | 90.70 | 92.41 | 93.79 | 93.09 | |
| BioBERT-Large | 82.59 | 86.73 | 84.52 | 87.27 | 91.24 | 89.21 | 90.58 | 93.28 | 91.91 | |
| BioBERT-Base | 85.02 | 88.11 | 86.51 | 89.69 | 92.24 | 90.95 | 92.50 | 94.13 | 93.31 | |
| BioM-ELECTRA | 84.90 | 88.56 | 86.55 | 89.80 | 92.36 | 91.06 | 92.62 | 94.16 | 93.38 | |
| BioELECTRA | 84.92 | 87.88 | 86.36 | 89.22 | 92.65 | 90.90 | 92.12 | 94.45 | 93.27 | |
| BioMed_RoBERTa | 84.36 | 89.25 | 86.67 | 87.90 | 93.82 | 90.76 | 91.52 | 95.31 | 93.38 | |
| LCEL¬ (Train _Aug + ASL) | Ensembled | 86.37 | 88.45 | 87.40 | 90.53 | 93.00 | 91.75 | 93.08 | 94.76 | 93.91 |
The overall comparison of data augmentation with different loss functions
| Policy . | Model . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|---|
| Train_Def + BCE | PubMedBERT | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| CovidBERT | 84.65 | 85.23 | 84.86 | 88.51 | 90.45 | 89.47 | 91.30 | 92.68 | 91.98 | |
| BioBERT-Large | 85.20 | 78.01 | 80.71 | 88.39 | 87.54 | 87.96 | 90.34 | 90.23 | 90.28 | |
| BioBERT-Base | 84.12 | 82.09 | 82.71 | 88.56 | 88.28 | 88.42 | 91.02 | 90.87 | 90.94 | |
| BioM-ELECTRA | 86.45 | 83.76 | 85.02 | 89.68 | 90.04 | 89.86 | 92.32 | 92.60 | 92.46 | |
| BioELECTRA | 83.34 | 85.77 | 84.48 | 88.59 | 90.81 | 89.68 | 91.68 | 93.09 | 92.38 | |
| BioMed_RoBERTa | 85.18 | 85.48 | 85.32 | 89.52 | 90.87 | 90.19 | 92.19 | 93.12 | 92.65 | |
| Train_Def + ASL | PubMedBERT | 85.49 | 86.33 | 85.90 | 89.70 | 91.24 | 90.47 | 92.43 | 93.43 | 92.93 |
| CovidBERT | 84.57 | 85.17 | 84.83 | 88.95 | 90.50 | 89.72 | 91.59 | 92.81 | 92.20 | |
| BioBERT-Large | 82.63 | 81.58 | 82.06 | 88.11 | 87.64 | 87.88 | 90.54 | 90.61 | 90.57 | |
| BioBERT-Base | 85.87 | 86.32 | 86.07 | 90.27 | 90.62 | 90.44 | 92.65 | 93.04 | 92.84 | |
| BioM-ELECTRA | 83.73 | 87.06 | 85.34 | 88.92 | 91.89 | 90.38 | 91.95 | 93.92 | 92.92 | |
| BioELECTRA | 83.67 | 87.43 | 85.47 | 88.61 | 91.49 | 90.03 | 91.70 | 93.50 | 92.59 | |
| BioMed_RoBERTa | 84.32 | 87.27 | 85.75 | 88.75 | 91.68 | 90.19 | 91.76 | 93.69 | 92.71 | |
| Train _Aug + BCE | PubMedBERT | 86.47 | 86.62 | 86.51 | 90.38 | 91.58 | 90.98 | 92.88 | 93.74 | 93.31 |
| CovidBERT | 85.26 | 86.62 | 85.62 | 89.74 | 91.36 | 90.54 | 92.45 | 93.48 | 92.96 | |
| BioBERT-Large | 85.88 | 81.02 | 83.33 | 90.54 | 87.56 | 89.03 | 92.13 | 90.64 | 91.38 | |
| BioBERT-Base | 88.16 | 85.14 | 86.45 | 90.98 | 91.46 | 91.22 | 93.04 | 93.49 | 93.26 | |
| BioM-ELECTRA | 87.94 | 85.39 | 86.52 | 90.70 | 91.24 | 90.97 | 92.88 | 93.43 | 93.15 | |
| BioELECTRA | 84.73 | 89.24 | 86.83 | 89.70 | 92.05 | 90.86 | 92.21 | 94.04 | 93.12 | |
| BioMed_RoBERTa | 89.10 | 83.62 | 86.11 | 91.76 | 90.21 | 90.98 | 93.40 | 92.54 | 92.97 | |
| Train _Aug + ASL | PubMedBERT | 84.86 | 87.93 | 86.36 | 89.75 | 92.45 | 91.08 | 92.54 | 94.34 | 93.43 |
| CovidBERT | 84.98 | 87.10 | 86.01 | 89.68 | 91.74 | 90.70 | 92.41 | 93.79 | 93.09 | |
| BioBERT-Large | 82.59 | 86.73 | 84.52 | 87.27 | 91.24 | 89.21 | 90.58 | 93.28 | 91.91 | |
| BioBERT-Base | 85.02 | 88.11 | 86.51 | 89.69 | 92.24 | 90.95 | 92.50 | 94.13 | 93.31 | |
| BioM-ELECTRA | 84.90 | 88.56 | 86.55 | 89.80 | 92.36 | 91.06 | 92.62 | 94.16 | 93.38 | |
| BioELECTRA | 84.92 | 87.88 | 86.36 | 89.22 | 92.65 | 90.90 | 92.12 | 94.45 | 93.27 | |
| BioMed_RoBERTa | 84.36 | 89.25 | 86.67 | 87.90 | 93.82 | 90.76 | 91.52 | 95.31 | 93.38 | |
| LCEL¬ (Train _Aug + ASL) | Ensembled | 86.37 | 88.45 | 87.40 | 90.53 | 93.00 | 91.75 | 93.08 | 94.76 | 93.91 |
| Policy . | Model . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|---|
| Train_Def + BCE | PubMedBERT | 84.71 | 87.25 | 85.93 | 89.24 | 90.96 | 90.09 | 92.02 | 93.20 | 92.61 |
| CovidBERT | 84.65 | 85.23 | 84.86 | 88.51 | 90.45 | 89.47 | 91.30 | 92.68 | 91.98 | |
| BioBERT-Large | 85.20 | 78.01 | 80.71 | 88.39 | 87.54 | 87.96 | 90.34 | 90.23 | 90.28 | |
| BioBERT-Base | 84.12 | 82.09 | 82.71 | 88.56 | 88.28 | 88.42 | 91.02 | 90.87 | 90.94 | |
| BioM-ELECTRA | 86.45 | 83.76 | 85.02 | 89.68 | 90.04 | 89.86 | 92.32 | 92.60 | 92.46 | |
| BioELECTRA | 83.34 | 85.77 | 84.48 | 88.59 | 90.81 | 89.68 | 91.68 | 93.09 | 92.38 | |
| BioMed_RoBERTa | 85.18 | 85.48 | 85.32 | 89.52 | 90.87 | 90.19 | 92.19 | 93.12 | 92.65 | |
| Train_Def + ASL | PubMedBERT | 85.49 | 86.33 | 85.90 | 89.70 | 91.24 | 90.47 | 92.43 | 93.43 | 92.93 |
| CovidBERT | 84.57 | 85.17 | 84.83 | 88.95 | 90.50 | 89.72 | 91.59 | 92.81 | 92.20 | |
| BioBERT-Large | 82.63 | 81.58 | 82.06 | 88.11 | 87.64 | 87.88 | 90.54 | 90.61 | 90.57 | |
| BioBERT-Base | 85.87 | 86.32 | 86.07 | 90.27 | 90.62 | 90.44 | 92.65 | 93.04 | 92.84 | |
| BioM-ELECTRA | 83.73 | 87.06 | 85.34 | 88.92 | 91.89 | 90.38 | 91.95 | 93.92 | 92.92 | |
| BioELECTRA | 83.67 | 87.43 | 85.47 | 88.61 | 91.49 | 90.03 | 91.70 | 93.50 | 92.59 | |
| BioMed_RoBERTa | 84.32 | 87.27 | 85.75 | 88.75 | 91.68 | 90.19 | 91.76 | 93.69 | 92.71 | |
| Train _Aug + BCE | PubMedBERT | 86.47 | 86.62 | 86.51 | 90.38 | 91.58 | 90.98 | 92.88 | 93.74 | 93.31 |
| CovidBERT | 85.26 | 86.62 | 85.62 | 89.74 | 91.36 | 90.54 | 92.45 | 93.48 | 92.96 | |
| BioBERT-Large | 85.88 | 81.02 | 83.33 | 90.54 | 87.56 | 89.03 | 92.13 | 90.64 | 91.38 | |
| BioBERT-Base | 88.16 | 85.14 | 86.45 | 90.98 | 91.46 | 91.22 | 93.04 | 93.49 | 93.26 | |
| BioM-ELECTRA | 87.94 | 85.39 | 86.52 | 90.70 | 91.24 | 90.97 | 92.88 | 93.43 | 93.15 | |
| BioELECTRA | 84.73 | 89.24 | 86.83 | 89.70 | 92.05 | 90.86 | 92.21 | 94.04 | 93.12 | |
| BioMed_RoBERTa | 89.10 | 83.62 | 86.11 | 91.76 | 90.21 | 90.98 | 93.40 | 92.54 | 92.97 | |
| Train _Aug + ASL | PubMedBERT | 84.86 | 87.93 | 86.36 | 89.75 | 92.45 | 91.08 | 92.54 | 94.34 | 93.43 |
| CovidBERT | 84.98 | 87.10 | 86.01 | 89.68 | 91.74 | 90.70 | 92.41 | 93.79 | 93.09 | |
| BioBERT-Large | 82.59 | 86.73 | 84.52 | 87.27 | 91.24 | 89.21 | 90.58 | 93.28 | 91.91 | |
| BioBERT-Base | 85.02 | 88.11 | 86.51 | 89.69 | 92.24 | 90.95 | 92.50 | 94.13 | 93.31 | |
| BioM-ELECTRA | 84.90 | 88.56 | 86.55 | 89.80 | 92.36 | 91.06 | 92.62 | 94.16 | 93.38 | |
| BioELECTRA | 84.92 | 87.88 | 86.36 | 89.22 | 92.65 | 90.90 | 92.12 | 94.45 | 93.27 | |
| BioMed_RoBERTa | 84.36 | 89.25 | 86.67 | 87.90 | 93.82 | 90.76 | 91.52 | 95.31 | 93.38 | |
| LCEL¬ (Train _Aug + ASL) | Ensembled | 86.37 | 88.45 | 87.40 | 90.53 | 93.00 | 91.75 | 93.08 | 94.76 | 93.91 |
In Table 6, Train_Def stands for the models trained only using the LitCovid training dataset, while Train_Aug means the models trained with data augmentation. It is worth noticing that the training policy of ‘Train_Def + BCE’ is identical to the previous work (13), which was one of the top-ranked systems during the LitCovid online competition. In contrast, ‘Train_Aug + ASL’ stands for the training policy proposed for LCEL.
As is shown in Table 6, for the training policy of ‘Train_Def + BCE’, it can be observed that all models have competitive performances with only slight differences due to their powerful feature representation abilities. This indicates that all pretrained models with biomedical knowledge can provide robust COVID-19–specific feature representations, which benefit the ultimate multi-label classification performance. In particular, PubMedBERT (28) acquires the highest MaF of 85.93%, while BioMed_RoBERTa (33) reports the best performance with an MiF of 90.19% and an EBF of 92.65%.
Moreover, compared with ‘Train_Def + BCE’, the training policy of ‘Train_Def + ASL’ consistently improves the performance of BioBERT-Base (30), BioM-ELECTRA (31), BioELECTRA (32) and BioMed-RoBERTa (33). This suggests that the ASL enables the models to decouple the impacts of positive and negative samples and helps the models focus more on the positive ones, which benefits the overall multi-label classification. Although there are some slight declines in the MaF of PubMedBERT (28) and CovidBERT (29), and the MiF of BioBERT-Large (30), the other F-measures of these pretrained models are still boosted due to ASL.
In contrast, when adopting data augmentation, the performance of the training policies based on Train_Aug significantly outperforms Train_Def. This indicates that inadequate training data make it difficult to learn the essential semantic representations, while data augmentation addresses the training deficiency effectively, which enables an overall improvement of the models. Regarding ‘Train_Aug + BCE’ and ‘Train_Aug + ASL’, both policies rival each other and exhibit competitive performance. This implies that a large number of external training data guarantee abundant priori semantic information, which provides a solid foundation for learning capability. Once adapted to the COVID-19 domain, the additional semantic information can help the pretrained models understand COVID-19–relevant topics better. After integrating all the fine-tuned pretrained models under the policy of ‘Train_Aug + ASL’, LCEL is able to obtain consistent superiority in all label-level and instance-based level F-measures, resulting in the highest MaF of 87.40%, MiF of 91.75% and EBF of 93.91%. In a word, the experimental results show the effectiveness of the proposed LCEL method, due to the efficient aggregation of multiple classifiers and ASL function.
System performance on the test dataset
In the following section, a comprehensive comparison between the state-of-the-art systems (13, 22–24, 39) and LCEL is performed on the BioCreative VII LitCovid test dataset. Since there were up to 80 different valid predictions submitted to the challenge (11) during the online competition, for a fair comparison, the organizers implemented an official baseline system that utilized a shallow embedding-based machine learning approach, namely ML-Net (39). Table 7 reports the official statistics of all submissions as well as the overall system comparison. The highest scores of F-measures are boldfaced in Table 7.
The comparison of system performance on the test dataset
| Team submission stats . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Mean | 86.70 | 80.12 | 81.91 | 89.67 | 86.24 | 87.78 | 89.85 | 88.87 | 89.31 |
| Std | 6.09 | 7.94 | 7.01 | 5.41 | 4.82 | 4.29 | 5.21 | 4.51 | 4.60 |
| Q1 | 84.63 | 75.45 | 76.51 | 88.03 | 84.52 | 85.41 | 86.99 | 86.19 | 86.68 |
| Median | 88.35 | 83.83 | 85.27 | 91.08 | 88.43 | 89.25 | 91.88 | 90.97 | 91.32 |
| Q3 | 90.79 | 85.55 | 86.70 | 92.51 | 89.64 | 90.83 | 93.53 | 91.92 | 92.54 |
| Baseline (ML-Net) | 83.64 | 73.09 | 76.55 | 87.56 | 81.42 | 84.37 | 88.49 | 85.14 | 86.78 |
| Bioformer | 90.38 | 88.23 | 88.75 | 93.67 | 90.02 | 91.81 | 94.14 | 92.56 | 93.34 |
| DUT914 | 87.78 | 88.30 | 87.60 | 91.34 | 92.17 | 91.75 | 93.50 | 94.38 | 93.94 |
| DonutNLP | 91.52 | 85.66 | 87.54 | 93.43 | 90.10 | 91.74 | 94.40 | 92.54 | 93.46 |
| PolyU_CBSNLP | 91.39 | 85.34 | 87.49 | 92.52 | 90.29 | 91.39 | 93.78 | 92.64 | 93.21 |
| LCEL | 89.79 | 92.53 | 90.94 | 91.38 | 94.75 | 93.03 | 93.49 | 96.09 | 94.77 |
| Team submission stats . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Mean | 86.70 | 80.12 | 81.91 | 89.67 | 86.24 | 87.78 | 89.85 | 88.87 | 89.31 |
| Std | 6.09 | 7.94 | 7.01 | 5.41 | 4.82 | 4.29 | 5.21 | 4.51 | 4.60 |
| Q1 | 84.63 | 75.45 | 76.51 | 88.03 | 84.52 | 85.41 | 86.99 | 86.19 | 86.68 |
| Median | 88.35 | 83.83 | 85.27 | 91.08 | 88.43 | 89.25 | 91.88 | 90.97 | 91.32 |
| Q3 | 90.79 | 85.55 | 86.70 | 92.51 | 89.64 | 90.83 | 93.53 | 91.92 | 92.54 |
| Baseline (ML-Net) | 83.64 | 73.09 | 76.55 | 87.56 | 81.42 | 84.37 | 88.49 | 85.14 | 86.78 |
| Bioformer | 90.38 | 88.23 | 88.75 | 93.67 | 90.02 | 91.81 | 94.14 | 92.56 | 93.34 |
| DUT914 | 87.78 | 88.30 | 87.60 | 91.34 | 92.17 | 91.75 | 93.50 | 94.38 | 93.94 |
| DonutNLP | 91.52 | 85.66 | 87.54 | 93.43 | 90.10 | 91.74 | 94.40 | 92.54 | 93.46 |
| PolyU_CBSNLP | 91.39 | 85.34 | 87.49 | 92.52 | 90.29 | 91.39 | 93.78 | 92.64 | 93.21 |
| LCEL | 89.79 | 92.53 | 90.94 | 91.38 | 94.75 | 93.03 | 93.49 | 96.09 | 94.77 |
The comparison of system performance on the test dataset
| Team submission stats . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Mean | 86.70 | 80.12 | 81.91 | 89.67 | 86.24 | 87.78 | 89.85 | 88.87 | 89.31 |
| Std | 6.09 | 7.94 | 7.01 | 5.41 | 4.82 | 4.29 | 5.21 | 4.51 | 4.60 |
| Q1 | 84.63 | 75.45 | 76.51 | 88.03 | 84.52 | 85.41 | 86.99 | 86.19 | 86.68 |
| Median | 88.35 | 83.83 | 85.27 | 91.08 | 88.43 | 89.25 | 91.88 | 90.97 | 91.32 |
| Q3 | 90.79 | 85.55 | 86.70 | 92.51 | 89.64 | 90.83 | 93.53 | 91.92 | 92.54 |
| Baseline (ML-Net) | 83.64 | 73.09 | 76.55 | 87.56 | 81.42 | 84.37 | 88.49 | 85.14 | 86.78 |
| Bioformer | 90.38 | 88.23 | 88.75 | 93.67 | 90.02 | 91.81 | 94.14 | 92.56 | 93.34 |
| DUT914 | 87.78 | 88.30 | 87.60 | 91.34 | 92.17 | 91.75 | 93.50 | 94.38 | 93.94 |
| DonutNLP | 91.52 | 85.66 | 87.54 | 93.43 | 90.10 | 91.74 | 94.40 | 92.54 | 93.46 |
| PolyU_CBSNLP | 91.39 | 85.34 | 87.49 | 92.52 | 90.29 | 91.39 | 93.78 | 92.64 | 93.21 |
| LCEL | 89.79 | 92.53 | 90.94 | 91.38 | 94.75 | 93.03 | 93.49 | 96.09 | 94.77 |
| Team submission stats . | MaP (%) . | MaR (%) . | MaF (%) . | MiP (%) . | MiR (%) . | MiF (%) . | EBP (%) . | EBR (%) . | EBF (%) . |
|---|---|---|---|---|---|---|---|---|---|
| Mean | 86.70 | 80.12 | 81.91 | 89.67 | 86.24 | 87.78 | 89.85 | 88.87 | 89.31 |
| Std | 6.09 | 7.94 | 7.01 | 5.41 | 4.82 | 4.29 | 5.21 | 4.51 | 4.60 |
| Q1 | 84.63 | 75.45 | 76.51 | 88.03 | 84.52 | 85.41 | 86.99 | 86.19 | 86.68 |
| Median | 88.35 | 83.83 | 85.27 | 91.08 | 88.43 | 89.25 | 91.88 | 90.97 | 91.32 |
| Q3 | 90.79 | 85.55 | 86.70 | 92.51 | 89.64 | 90.83 | 93.53 | 91.92 | 92.54 |
| Baseline (ML-Net) | 83.64 | 73.09 | 76.55 | 87.56 | 81.42 | 84.37 | 88.49 | 85.14 | 86.78 |
| Bioformer | 90.38 | 88.23 | 88.75 | 93.67 | 90.02 | 91.81 | 94.14 | 92.56 | 93.34 |
| DUT914 | 87.78 | 88.30 | 87.60 | 91.34 | 92.17 | 91.75 | 93.50 | 94.38 | 93.94 |
| DonutNLP | 91.52 | 85.66 | 87.54 | 93.43 | 90.10 | 91.74 | 94.40 | 92.54 | 93.46 |
| PolyU_CBSNLP | 91.39 | 85.34 | 87.49 | 92.52 | 90.29 | 91.39 | 93.78 | 92.64 | 93.21 |
| LCEL | 89.79 | 92.53 | 90.94 | 91.38 | 94.75 | 93.03 | 93.49 | 96.09 | 94.77 |
As shown in Table 7, the official baseline system ML-Net (39) reaches decent achievements with an MaF of 76.55%, an MiF of 84.37% and an EBF of 86.78%. The baseline performance is quite close to the Q1 statistics for all three F-measures, suggesting that ∼75% of the team submissions have more promising results than the official baseline method. In contrast, the average MaF, MiF and EBF of all submissions are as high as 81.91%, 87.78% and 89.31%, respectively, all of which are better than the baseline scores. However, although most submissions outperform the baseline system, there are still relatively large standard deviations among the submissions, with 7.01% to MaF, 4.82% to MiF and 4.60% to EBF, respectively.
Note that all four of the top-performing systems developed during the online competition, i.e. Bioformer (24), DonutNLP (23), DUT914 (22) and PolyU_CBSNLP (13), consistently achieved top-ranked performance in all three F-measures. Interestingly, all state-of-the-art systems (13, 22–24), more or less, adopted ensemble learning technologies. Specifically, Bioformer (24) investigated multiple pretrained models including PubMedBERT (28) and BioBERT (30) for the LitCovid multi-label classification problem. To enhance the representation abilities, Bioformer (24) further proposed to exploit a larger external dataset to fine-tune the pretrained models and achieved the best performance on MaF and MiF with scores of 88.75% and 91.81%, respectively. Similarly, DUT914 (22) proposed to merge the multiple feature representations from different pretrained models of CovidBERT (29) and BioBERT (30) to capture the crucial semantic clues for the LitCovid Track. Due to the feature enrichment, DUT914 (22) obtained the best EBF of 93.94%, surpassing Bioformer (24) by 0.6 units. DonutNLP (23) was another top-ranked system during the online competition, which utilized a voting-based ensemble learning method to integrate multiple BioBERT (30) models. Although DonutNLP (23) did not achieve a performance as high as Bioformer (24) and DUT914 (22), it still acquired a comparable performance of MaF, MiF and EBF, with scores of 87.54%, 91.74% and 93.46%, respectively. Likewise, our previous work PolyU_CBSNLP (13) also proposed to ensemble multiple pretrained models to tackle the challenging task. However, different from DonutNLP (23), the pretrained models with heterogeneous architectures were mainly taken into consideration in PolyU_CBSNLP (13), and in total, seven advanced pretrained models were adopted accordingly. Although our previous work (13) did not outperform the top systems, it still rivaled these systems and reached promising results with an MaF of 87.49%, an MiF of 91.39% and an EBF of 93.21%.
For the LCEL model, since there were seven different transformer-like pretrained models to be ensembled, during the training phase, only the models that performed the best on the development dataset were reserved for further integration. Compared with the above-mentioned state-of-the-art systems (13, 22–24, 39), LCEL consistently exhibits an overwhelming superiority in all F-measures, resulting in an MaF of 90.94, an MiF of 93.03 and an EBF of 94.77, respectively. Despite slightly lower precision, LCEL significantly improves the performance in all recall-based measures. This suggests the effectiveness of our proposed ensemble learning method. On the one hand, by adopting additional biomedical knowledge and data augmentation, LCEL is able to capture the supplementary semantic aspects related to COVID-19. On the other hand, benefiting from ASL, LCEL efficiently addresses the imbalanced label distribution, emphasizing more contributions to the positive samples. The experimental results illustrate the efficacy of the proposed ensemble learning method, which might lay the preliminary foundation for research in the COVID-19 domain.
Error analysis
To investigate the challenging issues in practice and provide insights for future work, we analyzed the errors in detail and grouped the main possible reasons as follows:
Implicit language expression
The proposed LCEL model aims to grasp critical semantic clues from literature contexts; however, in some cases, if there is a lack of such explicit evidence clearly expressed in the input texts, it would be difficult for LCEL to determine the final topics. For instance, in the article PMID:34202160, our LCEL cannot recognize the true topic of ‘Treatment’as there are no such explicit semantic clues to support that topic.
Contextual misunderstandings
Since the topic prediction of our proposed ensemble learning method largely relies on the contextual information provided by the input literature, sometimes, certain meaningful and remarkable words or phrases will result in misunderstandings of LCEL. For instance, in the article PMID:34291812, the main content of the literature describes treatments related to a COVID-19 infection; however, as the remarkably indicative word ‘prevention’ explicitly occurs in the title, our LCEL still outputs the label of ‘Prevention’ incorrectly.
Information deficiency
As pretrained models always impose constraints on the length of input texts, the overlong articles will be truncated before being fed into the downstream deep neural networks. However, some information would be inevitably discarded during this aggressive preprocessing. This in turn could cause unexpected difficulties for the LCEL model to recommend labels. In this study, around 17.8% of articles were truncated and 15.1% of the informative text is inevitably lost during the process due to the fixed sequence length of 512. For instance, during the preprocessing, the text of the article PMID:34227364 is shortened and some crucial information is dropped in the process, leading to the failure of recognizing the correct topic of ‘Mechanism’.
Predicting bias
Despite applying ASL to tackle the imbalanced label distribution problem, LCEL is still prone to pay more attention to the dominant topics aggressively while disregarding the tail ones conservatively. For instance, the article PMID:34205856 carries relatively short texts with no more than 30 words in its title and abstract. In this article, even though there is no such sign of ‘Treatment’, our LCEL model still recommends that topic, incorrectly.
Inconsistent annotation
In our experiments, some results show that parts of the false-positive COVID-19 topics identified by LCEL might be true and perhaps should be annotated in the LitCovid corpus. Taking the articles PMID:34338124 and PMID:34208057 into consideration, our LCEL model recommends the topic of ‘Case Report’ for both of them; however, even if some strongly indicative words (e.g. case, report, etc.) occur multiple times in the titles and the abstracts, the label is not annotated as the ground-truth answer. This is probably because of the inherent annotation disagreements of the LitCovid biocuration.
Conclusion and future work
This research proposed a novel ensemble learning of LCEL for COVID-19 multi-label classification, which integrated multiple powerful biomedical pretrained models. Specifically, seven advanced pretrained models with heterogeneous architectures were selected for ensemble learning. To enhance the representation abilities of deep neural models, additional biomedical knowledge and data augmentation strategies were exploited to fully utilize the semantic expressions. In light of the imbalanced label distribution, a novel ASL function was introduced to the LCEL model, which explicitly adjusted the negative–positive importance by assigning different exponential decay factors. Benefiting from ASL, the proposed model was able to dynamically decouple the modulations of the positive and negative samples during the training phase and focused more on the positive samples, while decreasing the contribution of negative ones. The experimental results on the LitCovid dataset achieved state-of-the-art performance, demonstrating the effectiveness of our proposed method.
Our research on the LitCovid dataset has exhibited promising results for the COVID-19 multi-label classification research. In future work, we will develop more advanced deep neural models with richer semantic features and sophisticated ensemble techniques to improve the current system for better performance.
Availability of data and materials
The datasets underlying this article are available in the BioCreative VII LitCovid Track at https://biocreative.bioinformatics.udel.edu/tasks/biocreative-vii/track-5/. The codes of the proposed LCEL model are available at https://github.com/JHnlp/LCEL.
Author contributions
J.G. and E.C. conceived the study; J.G. performed the data collection, training, prediction and analysis; J.G. and X.W. redesigned the experiment and data analysis; J.G., E.C., L.Q., G.Z. and C.H. co-wrote the paper and are responsible for various sections of theoretical interpretations, respectively. All authors contributed to the revised and approved the final manuscript.
Acknowledgements
Not applicable.
Funding
This research is supported by the research grants of The Hong Kong Polytechnic University Projects (#1-W182, #G-YW4H) and the National Natural Science Foundation of China (#61976147).
Conflict of interest.
None declared.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.