-
PDF
- Split View
-
Views
-
Cite
Cite
Daniel G. Jamieson, Martin Gerner, Farzaneh Sarafraz, Goran Nenadic, David L. Robertson, Towards semi-automated curation: using text mining to recreate the HIV-1, human protein interaction database, Database, Volume 2012, 2012, bas023, https://doi.org/10.1093/database/bas023
Close - Share Icon Share
Abstract
Manual curation has long been used for extracting key information found within the primary literature for input into biological databases. The human immunodeficiency virus type 1 (HIV-1), human protein interaction database (HHPID), for example, contains 2589 manually extracted interactions, linked to 14 312 mentions in 3090 articles. The advancement of text-mining (TM) techniques has offered a possibility to rapidly retrieve such data from large volumes of text to a high degree of accuracy. Here, we present a recreation of the HHPID using the current state of the art in TM. To retrieve interactions, we performed gene/protein named entity recognition (NER) and applied two molecular event extraction tools on all abstracts and titles cited in the HHPID. Our best NER scores for precision, recall and F-score were 87.5%, 90.0% and 88.6%, respectively, while event extraction achieved 76.4%, 84.2% and 80.1%, respectively. We demonstrate that over 50% of the HHPID interactions can be recreated from abstracts and titles. Furthermore, from 49 available open-access full-text articles, we extracted a total of 237 unique HIV-1–human interactions, as opposed to 187 interactions recorded in the HHPID from the same articles. On average, we extracted 23 times more mentions of interactions and events from a full-text article than from an abstract and title, with a 6-fold increase in the number of unique interactions. We further demonstrated that more frequently occurring interactions extracted by TM are more likely to be true positives. Overall, the results demonstrate that TM was able to recover a large proportion of interactions, many of which were found within the HHPID, making TM a useful assistant in the manual curation process. Finally, we also retrieved other types of interactions in the context of HIV-1 that are not currently present in the HHPID, thus, expanding the scope of this data set. All data is available at http://gnode1.mib.man.ac.uk/HIV1-text-mining.
Introduction
The human immunodeficiency virus type 1 (HIV-1), human protein interaction database (HHPID) is a manually curated database containing 2589 distinct HIV-1 to human protein interactions, linked to 14 312 mentions in 3090 Medline articles (1, 2). Each of these documented interactions is potentially of value to researchers studying HIV-1, where improved treatment strategies are in urgent demand for a disease that reported 33.3 million confirmed positive cases in 2009, leading to 1.8 million acquired immune deficiency syndrome-related deaths a year (3). As well as providing an instant resource to researchers seeking distinctive literature on specific HIV-1–human protein interactions, the HHPID has been used to construct detailed networks of the overall host–pathogen interactome (4) and has been vital in RNAi studies with HIV data (5–7).
The curation of the HHPID took over 7 years to complete and, ideally, it requires on-going updating. While an update based on manual curation is imminent, spanning from 2007 to 2011, future updates would benefit from some form of assisted curation effort. In the original design process of the HHPID, approximately 100 000 relevant HIV-1 documents were identified through PubMed queries, before further review and filtering reduced this number to 3200 (2). As of December 2011, a simple PubMed search for ‘HIV’ produces more than 233 000 results (including more than 64 000 new abstracts since 2007), highlighting the availability of a large body of potentially relevant literature for automated curation. Therefore, future updates to the HHPID will benefit from the ability to systematically process a much larger body of HIV-focused literature.
Text-mining (TM) techniques have emerged as a potential support solution to the knowledge extraction problem, helping to keep pace with the existing and ongoing expansion of primary literature. TM systems are designed to convert text data into manageable information and knowledge (8). Within TM, there exists a range of techniques used to identify, extract, analyse and visualize data stored within text (9). A large degree of focus within the field has been placed on accurately and exhaustively extracting molecular interactions (MIs) from biomedical text, supported by collaborative events such as the BioCreative and BioNLP shared tasks (10, 11). These have led to the overall advancement of biomedical TM, making large-scale data extraction an immediate possibility (12, 13). However, the quality of TM data has historically been scrutinized in comparison to manual curation, where aspects such as gene name ambiguity (14) and conflicting event relationships (15), have impeded its overall accuracy.
Existing forms of assisted curation using TM approaches have benefitted the manual curation process by reducing the scale and complexity of information that curators have to process. For example, Wiegers et al. (16) have demonstrated potential in ranking documents according to chemical, gene/protein and disease identifiers in text to augment the efficiency of manual curation of the Comparative Toxicogenomics Database. Another example comes from Kemper et al. (17) who have integrated TM components with a pathway visualizer and annotation tools to aid curators in generating metabolic and signalling pathways more effectively.
In this article, we explore the reconstruction of the HHPID using a suite of tailored state-of-the-art TM tools. The results and analyses demonstrate that TM is able to recover a large proportion of interactions found within the HHPID with reasonable recall and precision, in addition to expanding the scope of the database by identifying interactions between other types of entities. These techniques have demonstrated that future curation of the HHPID and indeed other MI databases can be assisted by TM helping speed up the curation process.
Methods
Figure 1 summarizes our approach for recreating and evaluating the HHPID using text mining tools. The method has four main steps: (i) text retrieval (using only citations from the HHPID), (ii) named-entity recognition (NER, finding mentions of molecules in text), (iii) molecular event extraction (finding any interactions that exist between entities) and (iv) various evaluations and comparisons of the results.
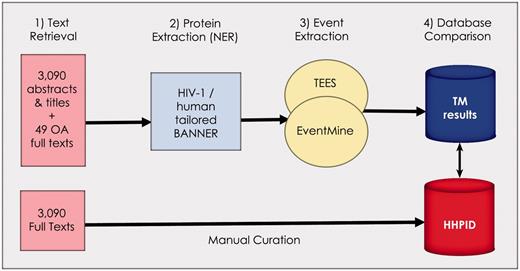
Summary of the methodology. Our methodology is divided into four stages: (1) retrieval of all abstracts and titles, as well as 49 open-access full texts from the 3090 citations in the HHPID, (2) proteins were extracted using an HIV-1/human tailored version of BANNER, (3) events were extracted using two event extraction tools (TEES and EventMine) and (4) a comparison of the results retrieved by TM was made with the manually curated HHPID.
Data
We limited our investigation to only those articles used in the HHPID to directly compare manual curation to TM. Of the 14 312 citations in the HHPID, we found 3090 of these to have unique PubMed identifiers (PMIDs). Only 49 articles (1.6%) were available through PubMed Central (PMC) as full-text open access (OA) articles. While it would be preferable to use full text for the entire set of 3090 cit-ations, the limited availability of OA articles restricted our main analysis to using abstracts and titles. To illustrate the value of using full-text articles, we also performed a separate experiment using the 49 OA articles.
Named entity recognition and normalization
To extract proteins from the text, we used BANNER, which has been ranked as one of the top performing NER systems by the BioCreative shared task III (18, 19). Since BANNER has been developed for use on NER across generic biomedical text, we decided to make adjustments to focus the tool on HIV-1 specific text so that we could enhance its overall performance (20). To identify any specific BANNER performance weaknesses on the HIV-related literature, we first evaluated the performance on a corpus of 50 randomly selected abstracts and titles from the HHPID (referred to as ‘Train-HIV’). We evaluated these abstracts using the same evaluation approach as used in NER evaluation in the BioCreative III shared task using precision, recall and F-score (10, 21).
The initial evaluation of BANNER revealed commonly occurring types of false positives such as protein regions (e.g. ‘V3’) or event mentions (e.g. ‘superoxide release’), and false negatives such as hyphenated entities (e.g. ‘tat-induced’) or entities contained within brackets (e.g. ‘(SOD1)’). While false positives were difficult to distinguish computationally, we were able to reduce the number of false negatives by providing an additional training data set with HIV-1–human interaction-specific classes of false negatives annotated in text. Furthermore, we designed and implemented post-processing modules to work in unison with BANNER and reduce false negatives by applying dictionaries of HIV-1 and top occurring human-related gene names to match untagged proteins from the text. We then evaluated our modified version of BANNER on a new corpus of 50 randomly selected abstracts and titles from the HHPID (referred to as ‘Test-HIV’).
In addition to recognition of gene names in text, we normalized our NER results to either HIV-1 or human genes using the Entrez Gene gene names, gene symbols and gene aliases (22). While normalization has traditionally been made difficult by intra- and inter-species gene name ambiguity (23), HIV-1’s small gene set (nine genes) and the knowledge that each document was HIV-1 relevant, helped us to more confidently and accurately associate genes with HIV-1. Gene names that could not be normalized to an HIV-1 dictionary were, wherever possible, mapped to a human dictionary. If they were not matched to either an HIV-1 dictionary or a human dictionary, they were classified as ‘other’.
Event extraction
We focus our investigation on specific types of events that represent interactions between proteins as defined by BioNLP’09 (11, 24). These interactions cover three types of protein metabolism (specifically, gene expression, transcription and protein catabolism), phosphorylation, localization, binding and regulatory events (regulation, positive regulation and negative regulation). Events are identified in text by using two event extraction tools, the Turku event extraction system (TEES) (25) and EventMine (15). The tools have been designed to conform to the BioNLP task. Events of gene expression, transcription, protein catabolism, phosphorylation and localization types are all required to act on a single gene or protein, called a theme. Binding events can have one or two gene/protein themes. Regulatory events differ in that their theme may be either a gene/protein or another event. While not required, a regulatory event can also have a gene/protein or another event as its cause. This allows for the possibility of ‘event chains’ involving multiple gene/proteins in multiple events. For example, the sentence “Tat increased the expression of NF kappa B” mentions an event chain that includes ‘expression of NF kappa B’ and positive regulation of that event by ‘Tat’ (Figure 2).
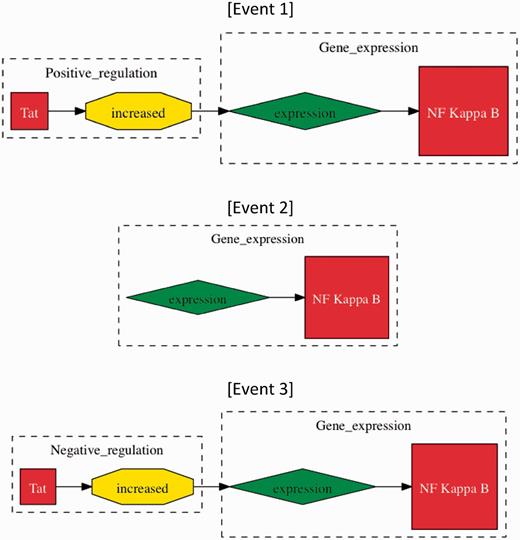
Methods of event evaluation. The three events have been extracted from the sentence “Tat increased the expression of NF kappa B”. In approximate evaluation, both events 1 and 2 would be counted as true positives, whereas only Event 1 would be considered a true positive in stringent event evaluation, as ‘Tat positive regulation (increased)’ is missing. Event 3 would be a false positive in both categories of evaluation, whereby ‘increased’ does not signify negative regulation.
We applied the two event extraction systems to 3090 titles and abstracts and 49 full-text articles associated with HHPID, after these had been tagged by the HIV-1/human tailored version of BANNER. We considered molecular events identified by either of the systems (union) or by both systems (intersection).
Event evaluation
Molecular interactions represented in the HHPID are characterized by 70 keywords that potentially indicate the type of interaction, many of which are potentially redundant (e.g. ‘binds’ and ‘complexes with’). To enable us to compare the event extraction results with interactions from the HHPID, we mapped 51 out of the 70 HHPID interaction keywords to the nine event types (see Supplementary File S1). The remaining 19 interaction keywords (such as ‘glycosylates’) were designated as ‘other’ in the results.
To assess the performance of the event extraction systems, we used our Test-HIV corpus of 50 abstracts and titles. Rather than evaluating single events as is commonplace in the BioNLP shared tasks (11), we evaluated ‘event chains’ since these represent a more complete depiction of the full interaction and have been represented as such in the HHPID. Event chains were evaluated under two different sets of rules: (i) Stringent event evaluation required that any recorded event chain should be represented in its entirety, i.e. without any falsely reported information in order to be classified as a true positive. (ii) Approximate event evaluation differs in that each reported event chain should be represented without any falsely reported information, although it may still be classified as a true positive if some information is missing. This allows for event chains with missing themes or causes to still be classified as true positives provided the rest of the captured data is correct. Figure 2 provides some examples of event evaluation methods.
Comparison of TM results to HHPID interactions
In order to ensure that any comparisons made between the TM results and the HHPID were fair, we firstly limited our analysis to only citations from the HHPID and interactions between HIV-1 and human molecules. When comparing interactions from the HHPID against TM, we used the Entrez gene IDs as specified in the database and cross-referenced TM entities with Entrez Gene HIV-1 and human gene names, gene symbols and gene synonyms.
It was not possible to automatically evaluate all TM-extracted interactions against the HHPID due to incompatibility of the data format representations (e.g. unspecified triggers, textual positions and full text/abstract origin of interactions within the HHPID). Instead, a random sample of 50 abstracts and titles from the data set was chosen and interactions reported within the HHPID as originating from the set were compared against those extracted through TM. We only considered interactions from the HHPID that were present within the abstracts and titles and not the full text. In addition, interactions that could not be extracted by TM, since they did not conform to the nine event types, but were present in the HHPID (e.g. ‘acetlyation’ interactions), were ignored.
A separate analysis was performed on the 49 PMC full-text OA articles that were cited in the HHPID. Following a similar procedure as above, we compared interactions retrieved from full text by TM against those retrieved from the same subset in the HHPID and those retrieved from only abstracts and titles by TM.
Results
We report two types of results: the generic accuracy of TM tools and accuracy specifically applied to the HHPID.
Accuracy of TM tools
The performance of the original version of BANNER (18) on our Test-HIV corpus showed precision, recall and F-score of 83.9%, 87.9% and 85.8%, respectively. When we used altered training data and combined BANNER with a post-processing module, our precision, recall and F-score were all improved to 87.5%, 90.0% and 88.6%, respectively, showing a marginal increase on the default BANNER configuration.
Table 1 shows the precision, recall and F-score for the event extraction tools. EventMine performed better than TEES in both stringent and approximate matching, with the highest precision, recall and F-score in approximate matching: 79.9%, 73.7% and 76.7%, respectively. When the results of both tools are merged in a union of events, recall and F-score are both notably higher in the stringent and approximate evaluations compared to individual tools and the precision is greater in the stringent evaluation. Our analysis showed that this was due to full event chains now being completely represented. However, the precision of the union is slightly lower (−3.5%) in the approximate matching. The highest precision is achieved in the intersection of the two tools (87.4%), although recall (46.2%) and F-score (60.4%) are considerably lower. We therefore decided to use the union of the two tools for further investigation.
Event extraction performance on the Test-HIV data set of 50 abstracts and titles
| . | Stringent . | Approximate . | ||||
|---|---|---|---|---|---|---|
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . | |
| TEES | 0.373 | 0.524 | 0.436 | 0.726 | 0.682 | 0.703 |
| EventMine | 0.460 | 0.622 | 0.529 | 0.799 | 0.737 | 0.767 |
| Union | 0.537 | 0.786 | 0.638 | 0.764 | 0.842 | 0.801 |
| Intersection | 0.663 | 0.392 | 0.493 | 0.874 | 0.462 | 0.604 |
| . | Stringent . | Approximate . | ||||
|---|---|---|---|---|---|---|
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . | |
| TEES | 0.373 | 0.524 | 0.436 | 0.726 | 0.682 | 0.703 |
| EventMine | 0.460 | 0.622 | 0.529 | 0.799 | 0.737 | 0.767 |
| Union | 0.537 | 0.786 | 0.638 | 0.764 | 0.842 | 0.801 |
| Intersection | 0.663 | 0.392 | 0.493 | 0.874 | 0.462 | 0.604 |
Event extraction performance on the Test-HIV data set of 50 abstracts and titles
| . | Stringent . | Approximate . | ||||
|---|---|---|---|---|---|---|
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . | |
| TEES | 0.373 | 0.524 | 0.436 | 0.726 | 0.682 | 0.703 |
| EventMine | 0.460 | 0.622 | 0.529 | 0.799 | 0.737 | 0.767 |
| Union | 0.537 | 0.786 | 0.638 | 0.764 | 0.842 | 0.801 |
| Intersection | 0.663 | 0.392 | 0.493 | 0.874 | 0.462 | 0.604 |
| . | Stringent . | Approximate . | ||||
|---|---|---|---|---|---|---|
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . | |
| TEES | 0.373 | 0.524 | 0.436 | 0.726 | 0.682 | 0.703 |
| EventMine | 0.460 | 0.622 | 0.529 | 0.799 | 0.737 | 0.767 |
| Union | 0.537 | 0.786 | 0.638 | 0.764 | 0.842 | 0.801 |
| Intersection | 0.663 | 0.392 | 0.493 | 0.874 | 0.462 | 0.604 |
Comparison of HIV-1–human interactions extracted by TM and the HHPID
Table 2 shows the total numbers of HIV-1–human molecular interactions for the HHPID and TM. We note that the TM results here are restricted to interactions between HIV-1 and human molecules only. The HHPID showed greater total numbers of interactions for all of the event types in comparison to TM. This is not surprising considering that the HHPID was derived from full text, whereas TM in this analysis was applied to abstracts and titles only. Table 3 further shows a comparison between the proteins involved in events (‘participants’) with the highest frequency in HIV-1–human interactions in the HHPID and TM. Here we observed eight out of ten of the same proteins shared between the two data sets.
The number of HIV-1–human interaction mentions extracted from 3090 citations: a comparison between the HHPID database and the TM results
| Interaction type . | Total HHIPD interactions (abstracts, titles and full text) . | Total TM interactions (abstracts and titles) . |
|---|---|---|
| Binding | 5534 | 1967 |
| Protein catabolism | 205 | 40 |
| Positive regulation | 3517 | 329 |
| Phosphorylation | 223 | 33 |
| Localization | 695 | 37 |
| Transcription | N/A | 31 |
| Regulation | 990 | 127 |
| Gene expression | N/A | 243 |
| Negative regulation | 1935 | 124 |
| Other | 518 | N/A |
| Total | 13 617 | 2931 |
| Interaction type . | Total HHIPD interactions (abstracts, titles and full text) . | Total TM interactions (abstracts and titles) . |
|---|---|---|
| Binding | 5534 | 1967 |
| Protein catabolism | 205 | 40 |
| Positive regulation | 3517 | 329 |
| Phosphorylation | 223 | 33 |
| Localization | 695 | 37 |
| Transcription | N/A | 31 |
| Regulation | 990 | 127 |
| Gene expression | N/A | 243 |
| Negative regulation | 1935 | 124 |
| Other | 518 | N/A |
| Total | 13 617 | 2931 |
The number of HIV-1–human interaction mentions extracted from 3090 citations: a comparison between the HHPID database and the TM results
| Interaction type . | Total HHIPD interactions (abstracts, titles and full text) . | Total TM interactions (abstracts and titles) . |
|---|---|---|
| Binding | 5534 | 1967 |
| Protein catabolism | 205 | 40 |
| Positive regulation | 3517 | 329 |
| Phosphorylation | 223 | 33 |
| Localization | 695 | 37 |
| Transcription | N/A | 31 |
| Regulation | 990 | 127 |
| Gene expression | N/A | 243 |
| Negative regulation | 1935 | 124 |
| Other | 518 | N/A |
| Total | 13 617 | 2931 |
| Interaction type . | Total HHIPD interactions (abstracts, titles and full text) . | Total TM interactions (abstracts and titles) . |
|---|---|---|
| Binding | 5534 | 1967 |
| Protein catabolism | 205 | 40 |
| Positive regulation | 3517 | 329 |
| Phosphorylation | 223 | 33 |
| Localization | 695 | 37 |
| Transcription | N/A | 31 |
| Regulation | 990 | 127 |
| Gene expression | N/A | 243 |
| Negative regulation | 1935 | 124 |
| Other | 518 | N/A |
| Total | 13 617 | 2931 |
Top 10 most frequent participants in events as presented in the HHPID and as extracted by TM
| Participant . | Total interactions . |
|---|---|
| HHPID | |
| Env gp160 | 4863 |
| Tat | 4247 |
| CD4 | 1188 |
| Vif | 1005 |
| Nef | 980 |
| Gag | 867 |
| Vpr | 790 |
| Gag-Pol | 541 |
| CXCR4 | 303 |
| CCR5 | 285 |
| Total interactions | 13 617 |
| TM | |
| Cd4 | 1290 |
| Tat | 1226 |
| Gp120 | 1161 |
| Nef | 531 |
| Env | 353 |
| Vpr | 230 |
| Cxcr4 | 230 |
| Cccr5 | 228 |
| Rev | 157 |
| Vpu | 65 |
| Total interactions | 2931 |
| Participant . | Total interactions . |
|---|---|
| HHPID | |
| Env gp160 | 4863 |
| Tat | 4247 |
| CD4 | 1188 |
| Vif | 1005 |
| Nef | 980 |
| Gag | 867 |
| Vpr | 790 |
| Gag-Pol | 541 |
| CXCR4 | 303 |
| CCR5 | 285 |
| Total interactions | 13 617 |
| TM | |
| Cd4 | 1290 |
| Tat | 1226 |
| Gp120 | 1161 |
| Nef | 531 |
| Env | 353 |
| Vpr | 230 |
| Cxcr4 | 230 |
| Cccr5 | 228 |
| Rev | 157 |
| Vpu | 65 |
| Total interactions | 2931 |
Top 10 most frequent participants in events as presented in the HHPID and as extracted by TM
| Participant . | Total interactions . |
|---|---|
| HHPID | |
| Env gp160 | 4863 |
| Tat | 4247 |
| CD4 | 1188 |
| Vif | 1005 |
| Nef | 980 |
| Gag | 867 |
| Vpr | 790 |
| Gag-Pol | 541 |
| CXCR4 | 303 |
| CCR5 | 285 |
| Total interactions | 13 617 |
| TM | |
| Cd4 | 1290 |
| Tat | 1226 |
| Gp120 | 1161 |
| Nef | 531 |
| Env | 353 |
| Vpr | 230 |
| Cxcr4 | 230 |
| Cccr5 | 228 |
| Rev | 157 |
| Vpu | 65 |
| Total interactions | 2931 |
| Participant . | Total interactions . |
|---|---|
| HHPID | |
| Env gp160 | 4863 |
| Tat | 4247 |
| CD4 | 1188 |
| Vif | 1005 |
| Nef | 980 |
| Gag | 867 |
| Vpr | 790 |
| Gag-Pol | 541 |
| CXCR4 | 303 |
| CCR5 | 285 |
| Total interactions | 13 617 |
| TM | |
| Cd4 | 1290 |
| Tat | 1226 |
| Gp120 | 1161 |
| Nef | 531 |
| Env | 353 |
| Vpr | 230 |
| Cxcr4 | 230 |
| Cccr5 | 228 |
| Rev | 157 |
| Vpu | 65 |
| Total interactions | 2931 |
To estimate how much of the HHPID we have replicated through TM, we compared interactions taken from abstracts and titles in the HHPID against HIV-1–human TM interactions over a set of 50 randomly selected citations from the HHPID. We were able to match 22 TM interactions to interactions within the HHPID, while 20 interactions that were present in the abstracts and titles were either missed or not fully extracted by TM. Thus, we estimate TM has recreated over 50% of interactions derived from the 3090 abstracts and titles within the HHPID without considering any potential data from full text. The value of using full text in TM is explored later in our analysis.
When we only considered frequently occurring unique HIV-1–human interactions, our results for TM were particularly encouraging. Table 4 shows the frequency of the top ten most commonly occurring HIV-1–human interactions extracted by TM. With our analysis restricted to unique interactions, TM achieves a similar number of total interactions (2069) in comparison to the HHPID (2589). All of the top 10 interactions retrieved automatically from text were true positives; however, only 7/10 were present within the HHPID. For example, ‘negative regulation of binding of gp120 to CD4’ is not present within the HHPID due to there being no regulation of binding interactions recorded within it. The ‘binding of gp120 to sCD4’ is not distinguished within the HHPID as an interaction, as CD4 is only recorded as ‘T-cell surface glycoprotein CD4 isoform 1 precursor’ and neglects the ‘soluble recombinant’ prefix of the CD4 nomenclature from the interaction. Instead, this information is presented within a reference sentence for the interaction in the HHPID and is unable to be filtered in a standard database query.
| TM interaction . | Frequency . | True positive . | Present in HHPID . |
|---|---|---|---|
| Binding of Gp120 to CD4 | 207 | Yes | Yes |
| Binding of Gp120 to CXCR4 | 32 | Yes | Yes |
| Binding of Tat to Cyclin T1 | 30 | Yes | Yes |
| Binding of Gp120 to CCR5 | 29 | Yes | Yes |
| Negative regulation of binding of Gp120 to CD4 | 24 | Yes | No |
| Binding of Vpu to CD4 | 19 | Yes | No |
| Binding of Gp120 to sCD4 | 18 | Yes | No |
| Binding of Nef to CD4 | 18 | Yes | Yes |
| Vpu positive regulation of protein catabolism of CD4 | 15 | Yes | Yes |
| Binding of Env to CD4 | 10 | Yes | Yes |
| Total unique mentions | 2069 | N/A | 2589 |
| TM interaction . | Frequency . | True positive . | Present in HHPID . |
|---|---|---|---|
| Binding of Gp120 to CD4 | 207 | Yes | Yes |
| Binding of Gp120 to CXCR4 | 32 | Yes | Yes |
| Binding of Tat to Cyclin T1 | 30 | Yes | Yes |
| Binding of Gp120 to CCR5 | 29 | Yes | Yes |
| Negative regulation of binding of Gp120 to CD4 | 24 | Yes | No |
| Binding of Vpu to CD4 | 19 | Yes | No |
| Binding of Gp120 to sCD4 | 18 | Yes | No |
| Binding of Nef to CD4 | 18 | Yes | Yes |
| Vpu positive regulation of protein catabolism of CD4 | 15 | Yes | Yes |
| Binding of Env to CD4 | 10 | Yes | Yes |
| Total unique mentions | 2069 | N/A | 2589 |
| TM interaction . | Frequency . | True positive . | Present in HHPID . |
|---|---|---|---|
| Binding of Gp120 to CD4 | 207 | Yes | Yes |
| Binding of Gp120 to CXCR4 | 32 | Yes | Yes |
| Binding of Tat to Cyclin T1 | 30 | Yes | Yes |
| Binding of Gp120 to CCR5 | 29 | Yes | Yes |
| Negative regulation of binding of Gp120 to CD4 | 24 | Yes | No |
| Binding of Vpu to CD4 | 19 | Yes | No |
| Binding of Gp120 to sCD4 | 18 | Yes | No |
| Binding of Nef to CD4 | 18 | Yes | Yes |
| Vpu positive regulation of protein catabolism of CD4 | 15 | Yes | Yes |
| Binding of Env to CD4 | 10 | Yes | Yes |
| Total unique mentions | 2069 | N/A | 2589 |
| TM interaction . | Frequency . | True positive . | Present in HHPID . |
|---|---|---|---|
| Binding of Gp120 to CD4 | 207 | Yes | Yes |
| Binding of Gp120 to CXCR4 | 32 | Yes | Yes |
| Binding of Tat to Cyclin T1 | 30 | Yes | Yes |
| Binding of Gp120 to CCR5 | 29 | Yes | Yes |
| Negative regulation of binding of Gp120 to CD4 | 24 | Yes | No |
| Binding of Vpu to CD4 | 19 | Yes | No |
| Binding of Gp120 to sCD4 | 18 | Yes | No |
| Binding of Nef to CD4 | 18 | Yes | Yes |
| Vpu positive regulation of protein catabolism of CD4 | 15 | Yes | Yes |
| Binding of Env to CD4 | 10 | Yes | Yes |
| Total unique mentions | 2069 | N/A | 2589 |
While these two instances of missing interactions from the HHPID can be accounted for by constraints in the way data in the HHPID is curated, there is no obvious reason as to why the ‘binding of Vpu to CD4’ is not present. We were able to confirm this interaction as a true positive from a number of references (26–29), all of which are present in the HHPID article set. We believe that—although binding of Vpu to CD4 has been documented as a direct interaction in a number of publications—the end result of this event is a down-regulation of CD4 and is documented in the HHPID as ‘Vpu degrades CD4’ and ‘Vpu downregulates CD4’—an interaction also qualified in the TM data set by ‘Vpu positive regulation of protein catabolism of CD4’. This discrepancy highlights issues for both the HHPID and TM. Here, it is evident that in the HHPID it is not completely clear from the interaction (when ignoring the reference sentence) that Vpu had bound to CD4 to cause its degradation. However, in TM, although both parts of the overall interaction (the binding and degradation) are represented in separate event chains, they cannot with the existing methodology be automatically linked together when spanning over one sentence. A combined TM and manual curation approach could help solve both of these problems, by using TM as a support to manual curation to provide additional descriptions for a candidate interaction.
Given the high number of binding events, we further analysed the most frequent interaction participants involved in this type of interaction. In Table 5, we compare the binding participants between the HHPID and TM for the HIV-1 Tat gene, as this gene was amongst the most frequent participants in both data sets. We observed similar numbers of total unique mentions of participants between the two data sets (388 for TM and 323 for the HHPID). ‘Cyclin T1’, ‘p-tefb’, ‘tbp’ and ‘Cyct1’ (a Cyclin T1 alias) were present in the top ten participants of both data sets. We observed ‘Sp1’ (11 mentions), ‘Pkr ’ (4 mentions) and ‘Puralpha’ (3 mentions) outside of the HHPID top ten, but within the top 10 in the TM results.
| Tat binding . | HHPID . | Tat binding . | TM . |
|---|---|---|---|
| P-Tefb | 57 | Tar | 51 |
| Cyclin T1 | 52 | Cyclin T1 | 30 |
| TBP | 22 | Tar RNA | 26 |
| CDK7 | 18 | p-tefb | 18 |
| CCNH | 17 | Tbp | 15 |
| ITGAV | 16 | Sp1 | 13 |
| ITGB3 | 16 | Pkr | 11 |
| CREBBP | 15 | Pp1 | 11 |
| GTF2H3 | 14 | Cyct1 | 9 |
| ERCC2 | 14 | Puralpha | 9 |
| Total interactants | 323 | Total interactants | 388 |
| Tat binding . | HHPID . | Tat binding . | TM . |
|---|---|---|---|
| P-Tefb | 57 | Tar | 51 |
| Cyclin T1 | 52 | Cyclin T1 | 30 |
| TBP | 22 | Tar RNA | 26 |
| CDK7 | 18 | p-tefb | 18 |
| CCNH | 17 | Tbp | 15 |
| ITGAV | 16 | Sp1 | 13 |
| ITGB3 | 16 | Pkr | 11 |
| CREBBP | 15 | Pp1 | 11 |
| GTF2H3 | 14 | Cyct1 | 9 |
| ERCC2 | 14 | Puralpha | 9 |
| Total interactants | 323 | Total interactants | 388 |
| Tat binding . | HHPID . | Tat binding . | TM . |
|---|---|---|---|
| P-Tefb | 57 | Tar | 51 |
| Cyclin T1 | 52 | Cyclin T1 | 30 |
| TBP | 22 | Tar RNA | 26 |
| CDK7 | 18 | p-tefb | 18 |
| CCNH | 17 | Tbp | 15 |
| ITGAV | 16 | Sp1 | 13 |
| ITGB3 | 16 | Pkr | 11 |
| CREBBP | 15 | Pp1 | 11 |
| GTF2H3 | 14 | Cyct1 | 9 |
| ERCC2 | 14 | Puralpha | 9 |
| Total interactants | 323 | Total interactants | 388 |
| Tat binding . | HHPID . | Tat binding . | TM . |
|---|---|---|---|
| P-Tefb | 57 | Tar | 51 |
| Cyclin T1 | 52 | Cyclin T1 | 30 |
| TBP | 22 | Tar RNA | 26 |
| CDK7 | 18 | p-tefb | 18 |
| CCNH | 17 | Tbp | 15 |
| ITGAV | 16 | Sp1 | 13 |
| ITGB3 | 16 | Pkr | 11 |
| CREBBP | 15 | Pp1 | 11 |
| GTF2H3 | 14 | Cyct1 | 9 |
| ERCC2 | 14 | Puralpha | 9 |
| Total interactants | 323 | Total interactants | 388 |
Other types of interactions retrieved by TM
As well as retrieving HIV-1–human molecular interactions, TM retrieved events and participants that were involved in other types of interactions or event chains. For example, in Table 5, the top occurring binding participant for Tat in the TM data set, Tar, was not present in the HHPID as this is an RNA molecule and the HHPID only contains protein–protein interactions.
Overall, TM retrieved 5674 events involving only a single HIV-1 protein, 7364 single human events, 437 HIV-1–HIV-1 interactions, 1265 human–human protein interactions and 243 interactions involving two or more participants (Table 6). Furthermore, we designated 8415 interactions as other, i.e. not involving an HIV-1 or human protein. We note that it is likely that this number is much lower, given that our normalization methods were not sufficient in categorizing all of the participants into their appropriate species.
Top 10 most frequently occurring participants within event chains in the TM results
| Interactant . | Number of interactions with . | Total interactions . | ||
|---|---|---|---|---|
| One participant . | Two interactants . | More than two interactants . | ||
| Cd4 | 1924 | 1290 | 62 | 3276 |
| Tat | 1244 | 1226 | 52 | 2522 |
| Gp120 | 1468 | 1161 | 60 | 2689 |
| Nef | 914 | 531 | 18 | 1463 |
| Env | 621 | 353 | 13 | 987 |
| Vpr | 301 | 230 | 6 | 537 |
| Cxcr4 | 357 | 230 | 15 | 602 |
| Cccr5 | 337 | 228 | 10 | 575 |
| Rev | 278 | 157 | 3 | 438 |
| Vpu | 184 | 65 | 5 | 254 |
| HIV-1 protein | 5674 | N/A | N/A | 5674 |
| Human protein | 7364 | N/A | N/A | 7364 |
| HIV-1–Human | N/A | 2931 | N/A | 2931 |
| HIV-1–HIV-1 | N/A | 437 | N/A | 437 |
| Human–Human | N/A | 1265 | N/A | 1265 |
| Other | 5560 | 2855 | N/A | 8415 |
| Total event chains | 18 598 | 7488 | 243 | 26 329 |
| Interactant . | Number of interactions with . | Total interactions . | ||
|---|---|---|---|---|
| One participant . | Two interactants . | More than two interactants . | ||
| Cd4 | 1924 | 1290 | 62 | 3276 |
| Tat | 1244 | 1226 | 52 | 2522 |
| Gp120 | 1468 | 1161 | 60 | 2689 |
| Nef | 914 | 531 | 18 | 1463 |
| Env | 621 | 353 | 13 | 987 |
| Vpr | 301 | 230 | 6 | 537 |
| Cxcr4 | 357 | 230 | 15 | 602 |
| Cccr5 | 337 | 228 | 10 | 575 |
| Rev | 278 | 157 | 3 | 438 |
| Vpu | 184 | 65 | 5 | 254 |
| HIV-1 protein | 5674 | N/A | N/A | 5674 |
| Human protein | 7364 | N/A | N/A | 7364 |
| HIV-1–Human | N/A | 2931 | N/A | 2931 |
| HIV-1–HIV-1 | N/A | 437 | N/A | 437 |
| Human–Human | N/A | 1265 | N/A | 1265 |
| Other | 5560 | 2855 | N/A | 8415 |
| Total event chains | 18 598 | 7488 | 243 | 26 329 |
The table presents the number of interactions with one, two or more interactants.
Top 10 most frequently occurring participants within event chains in the TM results
| Interactant . | Number of interactions with . | Total interactions . | ||
|---|---|---|---|---|
| One participant . | Two interactants . | More than two interactants . | ||
| Cd4 | 1924 | 1290 | 62 | 3276 |
| Tat | 1244 | 1226 | 52 | 2522 |
| Gp120 | 1468 | 1161 | 60 | 2689 |
| Nef | 914 | 531 | 18 | 1463 |
| Env | 621 | 353 | 13 | 987 |
| Vpr | 301 | 230 | 6 | 537 |
| Cxcr4 | 357 | 230 | 15 | 602 |
| Cccr5 | 337 | 228 | 10 | 575 |
| Rev | 278 | 157 | 3 | 438 |
| Vpu | 184 | 65 | 5 | 254 |
| HIV-1 protein | 5674 | N/A | N/A | 5674 |
| Human protein | 7364 | N/A | N/A | 7364 |
| HIV-1–Human | N/A | 2931 | N/A | 2931 |
| HIV-1–HIV-1 | N/A | 437 | N/A | 437 |
| Human–Human | N/A | 1265 | N/A | 1265 |
| Other | 5560 | 2855 | N/A | 8415 |
| Total event chains | 18 598 | 7488 | 243 | 26 329 |
| Interactant . | Number of interactions with . | Total interactions . | ||
|---|---|---|---|---|
| One participant . | Two interactants . | More than two interactants . | ||
| Cd4 | 1924 | 1290 | 62 | 3276 |
| Tat | 1244 | 1226 | 52 | 2522 |
| Gp120 | 1468 | 1161 | 60 | 2689 |
| Nef | 914 | 531 | 18 | 1463 |
| Env | 621 | 353 | 13 | 987 |
| Vpr | 301 | 230 | 6 | 537 |
| Cxcr4 | 357 | 230 | 15 | 602 |
| Cccr5 | 337 | 228 | 10 | 575 |
| Rev | 278 | 157 | 3 | 438 |
| Vpu | 184 | 65 | 5 | 254 |
| HIV-1 protein | 5674 | N/A | N/A | 5674 |
| Human protein | 7364 | N/A | N/A | 7364 |
| HIV-1–Human | N/A | 2931 | N/A | 2931 |
| HIV-1–HIV-1 | N/A | 437 | N/A | 437 |
| Human–Human | N/A | 1265 | N/A | 1265 |
| Other | 5560 | 2855 | N/A | 8415 |
| Total event chains | 18 598 | 7488 | 243 | 26 329 |
The table presents the number of interactions with one, two or more interactants.
Some of the most frequently occurring interactions that were not present in the HHPID, due to the restrictions in its scope are shown in Table 7. We noted that the majority of TM interactions that were false positives for HIV-1–HIV-1 and human–human MIs were each involving self-interactions, and as such can be filtered out easily. However, while these particular self-interactions represented false positives, we should take into account in future work that self-interactions may sometimes represent true positives as well (30).
| Interaction . | Interaction category . | Frequency . | True positive . |
|---|---|---|---|
| Binding of Tat to Tar | HIV-1–HIV-1 | 51 | Yes |
| Binding of tat to tat | HIV-1–HIV-1 | 21 | No |
| Binding of gp120 to gp41 | HIV-1–HIV-1 | 9 | Yes |
| Binding of gp120 to gp120 | HIV-1–HIV-1 | 8 | No |
| Binding of Nef to Nef | HIV-1–HIV-1 | 7 | No |
| Binding of CD4 to CD4 | Human–Human | 22 | No |
| Binding of CD4 to CXCR4 | Human–Human | 21 | Yes |
| Binding of CD4 to CCR5 | Human–Human | 16 | Yes |
| Binding of CCR5 to CCR5 | Human–Human | 5 | No |
| Binding of CCR5 to CXCR4 | Human–Human | 5 | No |
| Gp120 positive regulation of binding of CD4 to CD95 | More than 2 interactants | 2 | Yes |
| HIV-1 Tat positive regulation of HIV-1 Tat positive regulation of protein catabolism of iKappab | More than 2 interactants | 1 | No |
| P73 negative regulation of binding of Tat to Cyclin T1 | More than 2 interactants | 1 | Yes |
| Negative regulation of NF Kappa B/rel causes negative regulation of tat positive regulation of HIV-1 LTR | More than 2 interactants | 1 | Yes |
| Binding of CD4 to Okt4 antibody causes negative regulation of CD4 mobility | More than 2 interactants | 1 | Yes |
| Interaction . | Interaction category . | Frequency . | True positive . |
|---|---|---|---|
| Binding of Tat to Tar | HIV-1–HIV-1 | 51 | Yes |
| Binding of tat to tat | HIV-1–HIV-1 | 21 | No |
| Binding of gp120 to gp41 | HIV-1–HIV-1 | 9 | Yes |
| Binding of gp120 to gp120 | HIV-1–HIV-1 | 8 | No |
| Binding of Nef to Nef | HIV-1–HIV-1 | 7 | No |
| Binding of CD4 to CD4 | Human–Human | 22 | No |
| Binding of CD4 to CXCR4 | Human–Human | 21 | Yes |
| Binding of CD4 to CCR5 | Human–Human | 16 | Yes |
| Binding of CCR5 to CCR5 | Human–Human | 5 | No |
| Binding of CCR5 to CXCR4 | Human–Human | 5 | No |
| Gp120 positive regulation of binding of CD4 to CD95 | More than 2 interactants | 2 | Yes |
| HIV-1 Tat positive regulation of HIV-1 Tat positive regulation of protein catabolism of iKappab | More than 2 interactants | 1 | No |
| P73 negative regulation of binding of Tat to Cyclin T1 | More than 2 interactants | 1 | Yes |
| Negative regulation of NF Kappa B/rel causes negative regulation of tat positive regulation of HIV-1 LTR | More than 2 interactants | 1 | Yes |
| Binding of CD4 to Okt4 antibody causes negative regulation of CD4 mobility | More than 2 interactants | 1 | Yes |
| Interaction . | Interaction category . | Frequency . | True positive . |
|---|---|---|---|
| Binding of Tat to Tar | HIV-1–HIV-1 | 51 | Yes |
| Binding of tat to tat | HIV-1–HIV-1 | 21 | No |
| Binding of gp120 to gp41 | HIV-1–HIV-1 | 9 | Yes |
| Binding of gp120 to gp120 | HIV-1–HIV-1 | 8 | No |
| Binding of Nef to Nef | HIV-1–HIV-1 | 7 | No |
| Binding of CD4 to CD4 | Human–Human | 22 | No |
| Binding of CD4 to CXCR4 | Human–Human | 21 | Yes |
| Binding of CD4 to CCR5 | Human–Human | 16 | Yes |
| Binding of CCR5 to CCR5 | Human–Human | 5 | No |
| Binding of CCR5 to CXCR4 | Human–Human | 5 | No |
| Gp120 positive regulation of binding of CD4 to CD95 | More than 2 interactants | 2 | Yes |
| HIV-1 Tat positive regulation of HIV-1 Tat positive regulation of protein catabolism of iKappab | More than 2 interactants | 1 | No |
| P73 negative regulation of binding of Tat to Cyclin T1 | More than 2 interactants | 1 | Yes |
| Negative regulation of NF Kappa B/rel causes negative regulation of tat positive regulation of HIV-1 LTR | More than 2 interactants | 1 | Yes |
| Binding of CD4 to Okt4 antibody causes negative regulation of CD4 mobility | More than 2 interactants | 1 | Yes |
| Interaction . | Interaction category . | Frequency . | True positive . |
|---|---|---|---|
| Binding of Tat to Tar | HIV-1–HIV-1 | 51 | Yes |
| Binding of tat to tat | HIV-1–HIV-1 | 21 | No |
| Binding of gp120 to gp41 | HIV-1–HIV-1 | 9 | Yes |
| Binding of gp120 to gp120 | HIV-1–HIV-1 | 8 | No |
| Binding of Nef to Nef | HIV-1–HIV-1 | 7 | No |
| Binding of CD4 to CD4 | Human–Human | 22 | No |
| Binding of CD4 to CXCR4 | Human–Human | 21 | Yes |
| Binding of CD4 to CCR5 | Human–Human | 16 | Yes |
| Binding of CCR5 to CCR5 | Human–Human | 5 | No |
| Binding of CCR5 to CXCR4 | Human–Human | 5 | No |
| Gp120 positive regulation of binding of CD4 to CD95 | More than 2 interactants | 2 | Yes |
| HIV-1 Tat positive regulation of HIV-1 Tat positive regulation of protein catabolism of iKappab | More than 2 interactants | 1 | No |
| P73 negative regulation of binding of Tat to Cyclin T1 | More than 2 interactants | 1 | Yes |
| Negative regulation of NF Kappa B/rel causes negative regulation of tat positive regulation of HIV-1 LTR | More than 2 interactants | 1 | Yes |
| Binding of CD4 to Okt4 antibody causes negative regulation of CD4 mobility | More than 2 interactants | 1 | Yes |
Table 7 also shows that the HIV-1 trans-activation response element (TAR) is involved in Tat binding. It is interesting that this interaction was not present in the HHPID. Although a fundamental molecule involved in HIV-1’s biology (31), this TAR interaction is not included within the HHPID as it is an RNA molecule and the HHPID is limited to proteins only. This is also the case for the HIV-1 long-terminal repeat (LTR). To demonstrate the significance of TAR and LTR’s involvement within HIV-1 interactions, Table 8 shows their most frequently occurring interactions retrieved through TM and whether they are supported by the literature. Out of the 15 interactions involving LTR and TAR, only two were false positives.
| Interaction . | Frequency . | True positive . |
|---|---|---|
| Binding of Tat to TAR | 51 | Yes |
| Tat positive regulation of LTR | 11 | Yes |
| Binding of Cyclin T1 to TAR | 7 | No |
| Binding of RNA polymerase II to TAR | 6 | Yes |
| Negative regulation of binding of tat to TAR | 6 | Yes |
| Binding of CDK9 to TAR | 3 | No |
| Binding of TRP—185 to TAR | 3 | Yes |
| Binding of Tat to Cyclin T1 positive regulation of binding of Tat to TAR | 3 | Yes |
| Tat positive regulation of transcription of LTR | 3 | Yes |
| Binding of Tat to Vpr positive regulation of LTR | 2 | Yes |
| Tat positive regulation of tat positive regulation of LTR | 2 | Yes |
| Tat regulation of transcription LTR | 2 | Yes |
| Binding of LTR to SP1 | 2 | Yes |
| Vpr positive regulation of LTR | 2 | Yes |
| Ptb positive regulation of binding of RNA polymerase II to TAR | 2 | Yes |
| Interaction . | Frequency . | True positive . |
|---|---|---|
| Binding of Tat to TAR | 51 | Yes |
| Tat positive regulation of LTR | 11 | Yes |
| Binding of Cyclin T1 to TAR | 7 | No |
| Binding of RNA polymerase II to TAR | 6 | Yes |
| Negative regulation of binding of tat to TAR | 6 | Yes |
| Binding of CDK9 to TAR | 3 | No |
| Binding of TRP—185 to TAR | 3 | Yes |
| Binding of Tat to Cyclin T1 positive regulation of binding of Tat to TAR | 3 | Yes |
| Tat positive regulation of transcription of LTR | 3 | Yes |
| Binding of Tat to Vpr positive regulation of LTR | 2 | Yes |
| Tat positive regulation of tat positive regulation of LTR | 2 | Yes |
| Tat regulation of transcription LTR | 2 | Yes |
| Binding of LTR to SP1 | 2 | Yes |
| Vpr positive regulation of LTR | 2 | Yes |
| Ptb positive regulation of binding of RNA polymerase II to TAR | 2 | Yes |
| Interaction . | Frequency . | True positive . |
|---|---|---|
| Binding of Tat to TAR | 51 | Yes |
| Tat positive regulation of LTR | 11 | Yes |
| Binding of Cyclin T1 to TAR | 7 | No |
| Binding of RNA polymerase II to TAR | 6 | Yes |
| Negative regulation of binding of tat to TAR | 6 | Yes |
| Binding of CDK9 to TAR | 3 | No |
| Binding of TRP—185 to TAR | 3 | Yes |
| Binding of Tat to Cyclin T1 positive regulation of binding of Tat to TAR | 3 | Yes |
| Tat positive regulation of transcription of LTR | 3 | Yes |
| Binding of Tat to Vpr positive regulation of LTR | 2 | Yes |
| Tat positive regulation of tat positive regulation of LTR | 2 | Yes |
| Tat regulation of transcription LTR | 2 | Yes |
| Binding of LTR to SP1 | 2 | Yes |
| Vpr positive regulation of LTR | 2 | Yes |
| Ptb positive regulation of binding of RNA polymerase II to TAR | 2 | Yes |
| Interaction . | Frequency . | True positive . |
|---|---|---|
| Binding of Tat to TAR | 51 | Yes |
| Tat positive regulation of LTR | 11 | Yes |
| Binding of Cyclin T1 to TAR | 7 | No |
| Binding of RNA polymerase II to TAR | 6 | Yes |
| Negative regulation of binding of tat to TAR | 6 | Yes |
| Binding of CDK9 to TAR | 3 | No |
| Binding of TRP—185 to TAR | 3 | Yes |
| Binding of Tat to Cyclin T1 positive regulation of binding of Tat to TAR | 3 | Yes |
| Tat positive regulation of transcription of LTR | 3 | Yes |
| Binding of Tat to Vpr positive regulation of LTR | 2 | Yes |
| Tat positive regulation of tat positive regulation of LTR | 2 | Yes |
| Tat regulation of transcription LTR | 2 | Yes |
| Binding of LTR to SP1 | 2 | Yes |
| Vpr positive regulation of LTR | 2 | Yes |
| Ptb positive regulation of binding of RNA polymerase II to TAR | 2 | Yes |
Full-text TM analysis
Table 9 shows most frequent interactions extracted from the 49 articles cited within the HHPID which were open access and available for text mining. We compared HIV-1–human interactions extracted from full text, abstracts and titles and those denoted within the HHPID for this set of articles. For the top 10 interactions retrieved through TM applied on full text, we could only account for four in the HHPID, despite all 10 being true positives, indicating that potentially 60% of top-ranked full-text TM interactions might be missing from the HHPID. In total, there were 237 unique HIV-1–human interactions extracted from the 49 articles. This is 27% more than what is in the HHPID from the same subset, suggesting a potential gap in the interaction references in the HHPID. Although TM will have almost certainly reported some false positives (and false negatives for that matter) within these, the absence of 6 out of 10 true positive interactions found by full-text TM suggests that manual curation is not as exhaustive as we may have come to expect.
Top 10 most frequent interactions retrieved from 49 OA full-text articles with TM
| Interaction . | Full text TM frequency . | Abstracts and titles TM frequency . | HHPID frequency . | True Positive . |
|---|---|---|---|---|
| Binding of Vif to APOBEC3G | 27 | 0 | No | TP |
| Binding of DC-SIGN to gp120 | 22 | 0 | Yes | TP |
| Binding of Nef to ABCA1 | 20 | 1 | No | TP |
| Binding of gp120 to CD4 | 17 | 0 | Yes | TP |
| Nef Positive regulation of Rac | 16 | 2 | No | TP |
| Binding of Tat to CDK2 | 15 | 1 | No | TP |
| Binding of DOCK2 to Nef | 14 | 0 | Yes | TP |
| Binding of Nef to ELMO1 | 14 | 0 | Yes | TP |
| Vif Positive regulation of protein catabolism of APOBEC3G | 13 | 0 | No | TP |
| Binding of gp120 to CXCR4 | 11 | 0 | 0 | TP |
| Total unique HIV-1–human interactions | 237 | 39 | 187 | N/A |
| Total HIV-1–human interaction mentions | 4342 | 40 | N/A | N/A |
| Other mentions (single events, HIV-1–HIV-1 interactions, etc.) | 6995 | 441 | N/A | N/A |
| Total mentions | 11 337 | 481 | N/A | N/A |
| Interaction . | Full text TM frequency . | Abstracts and titles TM frequency . | HHPID frequency . | True Positive . |
|---|---|---|---|---|
| Binding of Vif to APOBEC3G | 27 | 0 | No | TP |
| Binding of DC-SIGN to gp120 | 22 | 0 | Yes | TP |
| Binding of Nef to ABCA1 | 20 | 1 | No | TP |
| Binding of gp120 to CD4 | 17 | 0 | Yes | TP |
| Nef Positive regulation of Rac | 16 | 2 | No | TP |
| Binding of Tat to CDK2 | 15 | 1 | No | TP |
| Binding of DOCK2 to Nef | 14 | 0 | Yes | TP |
| Binding of Nef to ELMO1 | 14 | 0 | Yes | TP |
| Vif Positive regulation of protein catabolism of APOBEC3G | 13 | 0 | No | TP |
| Binding of gp120 to CXCR4 | 11 | 0 | 0 | TP |
| Total unique HIV-1–human interactions | 237 | 39 | 187 | N/A |
| Total HIV-1–human interaction mentions | 4342 | 40 | N/A | N/A |
| Other mentions (single events, HIV-1–HIV-1 interactions, etc.) | 6995 | 441 | N/A | N/A |
| Total mentions | 11 337 | 481 | N/A | N/A |
Top 10 most frequent interactions retrieved from 49 OA full-text articles with TM
| Interaction . | Full text TM frequency . | Abstracts and titles TM frequency . | HHPID frequency . | True Positive . |
|---|---|---|---|---|
| Binding of Vif to APOBEC3G | 27 | 0 | No | TP |
| Binding of DC-SIGN to gp120 | 22 | 0 | Yes | TP |
| Binding of Nef to ABCA1 | 20 | 1 | No | TP |
| Binding of gp120 to CD4 | 17 | 0 | Yes | TP |
| Nef Positive regulation of Rac | 16 | 2 | No | TP |
| Binding of Tat to CDK2 | 15 | 1 | No | TP |
| Binding of DOCK2 to Nef | 14 | 0 | Yes | TP |
| Binding of Nef to ELMO1 | 14 | 0 | Yes | TP |
| Vif Positive regulation of protein catabolism of APOBEC3G | 13 | 0 | No | TP |
| Binding of gp120 to CXCR4 | 11 | 0 | 0 | TP |
| Total unique HIV-1–human interactions | 237 | 39 | 187 | N/A |
| Total HIV-1–human interaction mentions | 4342 | 40 | N/A | N/A |
| Other mentions (single events, HIV-1–HIV-1 interactions, etc.) | 6995 | 441 | N/A | N/A |
| Total mentions | 11 337 | 481 | N/A | N/A |
| Interaction . | Full text TM frequency . | Abstracts and titles TM frequency . | HHPID frequency . | True Positive . |
|---|---|---|---|---|
| Binding of Vif to APOBEC3G | 27 | 0 | No | TP |
| Binding of DC-SIGN to gp120 | 22 | 0 | Yes | TP |
| Binding of Nef to ABCA1 | 20 | 1 | No | TP |
| Binding of gp120 to CD4 | 17 | 0 | Yes | TP |
| Nef Positive regulation of Rac | 16 | 2 | No | TP |
| Binding of Tat to CDK2 | 15 | 1 | No | TP |
| Binding of DOCK2 to Nef | 14 | 0 | Yes | TP |
| Binding of Nef to ELMO1 | 14 | 0 | Yes | TP |
| Vif Positive regulation of protein catabolism of APOBEC3G | 13 | 0 | No | TP |
| Binding of gp120 to CXCR4 | 11 | 0 | 0 | TP |
| Total unique HIV-1–human interactions | 237 | 39 | 187 | N/A |
| Total HIV-1–human interaction mentions | 4342 | 40 | N/A | N/A |
| Other mentions (single events, HIV-1–HIV-1 interactions, etc.) | 6995 | 441 | N/A | N/A |
| Total mentions | 11 337 | 481 | N/A | N/A |
A comparison of HIV-1–human interactions extracted from full-text to those extracted using only abstracts and titles revealed over a 6-fold increase in the number of unique interactions. Only three of the top 10 interactions from full-text TM were found in the abstracts and titles TM subset. Overall, TM on full text recorded an average of 231 interaction or single event mentions per article in contrast to just 10 in abstracts and titles, an increase of 23 times. These results provide a compelling justification for the use of full text as opposed to only abstracts and titles in TM.
Discussion
Our custom BANNER system was able to achieve precision, recall and F-score of 88%, 90% and 89%, respectively using a modified, specially tailored training data set and a post-processing module utilizing a dictionary with HIV-1 and top occurring human genes. Although only marginally better than the original system, these scores demonstrated TM to be capable of extracting genes and gene products from HIV text to a useful level. An error analysis shows that commonly occurring false positives were acronyms such as cell line names (e.g. HeLa) or strain names (e.g. HIV-1 subtype B).
For event evaluation, we chose to use a union of two event extraction tools, which—under our most strict method of evaluation—showed precision, recall and F-score of 54%, 79% and 64%, respectively. Our approximate form of event evaluation for our best system showed precision, recall and F score of 76%, 84% and 80%, respectively. These results indicate that a large proportion of false positives from our stringent evaluation were caused not through falsely reported information, but through incomplete event chains, such as missing interaction causes or binding partners. Here, there is potential to improve on the performance of event extraction through completing the event chains that have missing information. However, generally the greatest challenge for event extraction tools comes from apprehending the various writing styles employed by different authors. False positive events were most persistently caused by complex grammatical sentences or just poor grammar, making it difficult for automated tools to ascertain their intended meaning. Figure 3 provides some examples of typical false positives.
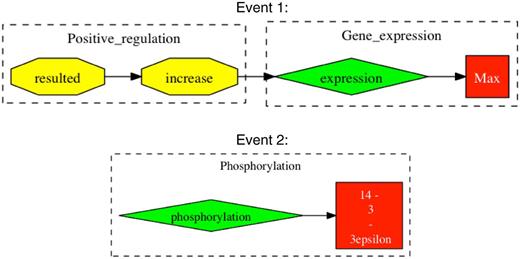
Examples of falsely reported event chains. Events are extracted from the sentence “In parallel to the modulation of cell growth, gp 120 at low concentrations resulted in an increase in the expression of c-Myc, Max, and 14–3-3epsilon proteins and phosphorylation of ATP-dependent tyrosine kinases (Akt) at Ser (473)”. Taken from Ref. (20). Event 1 shows an example of an incomplete event chain, where gp120 is missing as the cause for positive regulation. In Event 2, there is falsely reported information in that 14-3-3epsilon is expressed and not phosphorylated.
TM versus manual curation
We have successfully managed to recreate a large proportion of the interactions denoted within the HHPID using the current state of the art in TM. We have shown that TM tools are at least capable of precisely replicating over 50% of the interactions denoted within the HHPID from an evaluation sample of 50 abstracts and titles. Considering the manual curation of the HHPID took 7 years to perform, our tools have proven to be markedly more efficient by replicating a large percentage of this data automatically in a matter of hours.
Across the full list of citations within the HHPID, we have retrieved 2069 total unique HIV-1–human interaction mentions in comparison to 2589 unique HHPID interactions. Although some of these TM interactions probably represent false positives, this result is still extremely encouraging considering that curators of the HHPID had access to interactions from full text as well as abstracts and titles. From these HIV-1–human interactions, we found 7 of the top 10 binding interactants between Tat retrieved by TM to be present in the HHPID. Thus, we feel that those interactions recovered using TM represent a strong demonstration of how manual curation could be supported by sophisticated TM.
A top participant detected by TM for Tat binding that was not present in the HHPID was the HIV-1 TAR element. We found that the HHPID does not have any mentions of the HIV-1 TAR or any other RNA interactions involving HIV. It was not an objective of the HHPID to document these kinds of interactions, although, they are a potentially valuable resource to researchers studying HIV-1. To determine the role of TAR and another HIV-1 RNA molecule, the LTR, we highlighted interactions involving only these molecules (Table 8). Across the 15 interactions that we examined, only two were false positives and thus, we feel TM have the potential to identify valuable information from HIV-specific text on HIV-1 interactions that are not currently present in the HHPID. Given the other types of interactions that could be extracted (interactions between HIV-1 molecules, interactions between human molecules, interactions between two or more participants, etc.), TM tools could facilitate a semi-automated approach to the expansion of the scope of the HHPID database.
From five interactions involving more than two participants that we examined (Table 7), we were able to find four true positives. The true positives for interactions involving more than two participants are especially beneficial as that they provide a more complete illustration of interactions in contrast to the HHPID. Figure 4 shows an example.
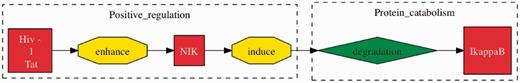
TM interaction involving two or more participants. This event was extracted from the sentence “HIV-1 Tat can substantially enhance the capacity of NIK to induce IkappaB degradation” (32). Here, we can see that the full interaction is identified by TM, across multiple participants and events. The HHPID documents this same interaction as ‘Tat enhances mitogen-activated protein kinase kinase kinase 14’, which is clearly a misrepresentation of the actual full interaction.
To consider the potential of full-text TM, we investigated the available OA articles cited in the HHPID. Interactions extracted from this subset highlighted that 6 of the top 10 interactions retrieved by full-text TM were missing, with 27% fewer unique interactions compared to the HHPID. These particular full-text articles referred to large numbers of gene and gene product mentions, contributing to some 11 337 interaction mentions as deduced by TM. While inaccuracies of TM cannot be ignored, these results do perhaps draw attention to limitations of manual curation, especially when dealing with more interaction-saturated literature, e.g. in high-throughput studies which are likely to contain more interaction mentions. However, it should be noted that curators from the HHPID may have chosen to only document the most important interactions denoted within these papers, accounting for the lower numbers of interactions.
In our subset of OA full-text articles, a comparison of TM using only abstracts and titles of the same articles exposed a significantly lower frequency of interaction mentions. On average, there were only 10 interaction mentions in abstracts and titles in contrast to 231 in full text. When only unique HIV-1–human interaction mentions were considered, full text still showed a 6-fold increase in data, with seven of the top 10 full-text TM interactions not present in the abstracts and titles TM data set. Although it has already been demonstrated that full text contains more information (33), only a small number of more than 233 000 HIV-related articles are accessible through PMC OA, thus, limiting the full potential of full-text TM to provide a large-scale systematic approach to information extraction from the entire literature.
One major weakness in our approach was the lack of an advanced normalization system able to fully categorize all of our retrieved participants into either HIV-1 or human species types. The dictionary-based methods we used can potentially be improved by using more sophisticated normalization systems such as GNAT (34, 35) or GeneTUKit (34, 36). Better normalization of participants will enable us to more precisely identify the interactions that TM has retrieved. However, we will be careful to ensure that useful context in descriptive prefixes and suffixes of molecules, e.g. ‘mutant’, are not lost while normalizing, as this information can potentially be useful to researchers in understanding what was originally documented.
Conclusion
In this article, we explored the potential of a TM-driven approach to curation of the HHPID. The results and analyses demonstrate that TM is able to recover a large proportion of interactions found within the HHPID with a reasonable recall/precision ratio, in addition to potentially expanding the scope of the database by identifying interactions between other types of entities. In principle, TM methods are more likely to retrieve true positives that are more frequently recorded in the literature. With such a large body of citations available for HIV, we believe that in the future we will be able to apply confidence to interactions based on how frequently they were recorded, and thus provide better support to the curation process.
Our analysis of full-text TM has revealed a convincing support for its usefulness, compared to solitary abstracts and titles. With such a dramatic difference in the frequencies of interaction mentions, we believe that in our future work we will be able to retrieve huge numbers of interactions if we have access to all full-text articles. A potential problem in full-text analysis in comparison to using only abstracts and titles will be to identify the ‘value’ and ‘novelty’ of an interaction, where aspects such as defining interactions as ‘referenced’ or ‘recorded’ will present new TM challenges. However, we believe neglecting such huge amounts of potentially valuable data would vastly hinder any future efforts to curate a more complete HIV-1–human protein interaction database.
Overall, although it is unlikely that TM will ever be able to replicate the accuracy that manual curation can achieve in MI extraction, its main strength is in the speed at which it can generate data that can be used to, amongst other aspects, support the curation process. Our results have shown that TM can retrieve reasonably accurate results for MI extraction and therefore a TM-assisted manual curation approach could be most beneficial, in particular for the more frequent interactions that can be checked first via references to the text. In the future, we intend to apply the current techniques with any improvements to the full list of HIV-1 citations in Medline and PMC, and make our results available to researchers online. The corpora generated are available on request.
Funding
Biotechnology and Biological Sciences Research Council (BBSRC) CASE studentship with industry partner Pfizer (BB/H016694/1 to D.G.J.); University of Manchester and a BBSRC CASE studentship with industry partner BioMed Central (to M.G.). Funding for open access charge: BB/H016694/1.
Conflict of interest. None declared.
The new version of the manuscript includes corrections to misspelled mentions of a software and an updated citation and acknowledgment.
Acknowledgements
The authors would like to thank Ben Sidders from Pfizer, UK for helpful comments and feedback; and Jonathan Dickerson and Jamie MacPherson for providing help throughout the investigation. We thank Makoto Miwa (the National Centre for Text Mining, University of Manchester) for advance access and help with EventMine (http://www.nactem.ac.uk/EventMine/) and Jari Björne (University of Turku) for help with TEES (http://bionlp.utu.fi/eventextractionsoftware.html). We would also like to thank Roger Ptak and William Fu for feedback and comments on the curation of the HHPID.



