-
PDF
- Split View
-
Views
-
Cite
Cite
Allan Peter Davis, Robin J. Johnson, Kelley Lennon-Hopkins, Daniela Sciaky, Michael C. Rosenstein, Thomas C. Wiegers, Carolyn J. Mattingly, Targeted journal curation as a method to improve data currency at the Comparative Toxicogenomics Database, Database, Volume 2012, 2012, bas051, https://doi.org/10.1093/database/bas051
Close - Share Icon Share
Abstract
The Comparative Toxicogenomics Database (CTD) is a public resource that promotes understanding about the effects of environmental chemicals on human health. CTD biocurators read the scientific literature and manually curate a triad of chemical–gene, chemical–disease and gene–disease interactions. Typically, articles for CTD are selected using a chemical-centric approach by querying PubMed to retrieve a corpus containing the chemical of interest. Although this technique ensures adequate coverage of knowledge about the chemical (i.e. data completeness), it does not necessarily reflect the most current state of all toxicological research in the community at large (i.e. data currency). Keeping databases current with the most recent scientific results, as well as providing a rich historical background from legacy articles, is a challenging process. To address this issue of data currency, CTD designed and tested a journal-centric approach of curation to complement our chemical-centric method. We first identified priority journals based on defined criteria. Next, over 7 weeks, three biocurators reviewed 2425 articles from three consecutive years (2009–2011) of three targeted journals. From this corpus, 1252 articles contained relevant data for CTD and 52 752 interactions were manually curated. Here, we describe our journal selection process, two methods of document delivery for the biocurators and the analysis of the resulting curation metrics, including data currency, and both intra-journal and inter-journal comparisons of research topics. Based on our results, we expect that curation by select journals can (i) be easily incorporated into the curation pipeline to complement our chemical-centric approach; (ii) build content more evenly for chemicals, genes and diseases in CTD (rather than biasing data by chemicals-of-interest); (iii) reflect developing areas in environmental health and (iv) improve overall data currency for chemicals, genes and diseases.
Database URL: http://ctdbase.org/
Introduction
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org) is a publicly available research tool that helps investigators understand the connections between environmental chemicals and gene products, and their potential effects on human health (1–4). CTD biocurators read the scientific literature and manually curate a triad of core data describing chemical–gene, chemical–disease and gene–disease interactions (5). Although manually curated databases provide a rich source of reliable information, they face challenges with respect to keeping data current and complete (6). At CTD, ‘data currency’ refers to how up-to-date (current) the information is in the database and ‘data completeness’ refers to how comprehensive the information is about a particular chemical, gene or disease. These two concepts, while not mutually exclusive, need to be balanced with respect to prioritizing and selecting what articles are to be manually curated.
Based on use studies, CTD prioritizes curation using a chemical-centric approach. Since 2005, we have maintained and updated a Chemical Priority Matrix of over 2400 compounds that are of research interest from seven independent sources, including three government toxicology programs, three collaborative groups and our users (5). This matrix has been used as a guide for prioritizing and selecting the scientific literature and has resulted in the manual curation of 34 084 articles to form the core of CTD. This chemical-centric approach helps ensure data completeness for any individual chemical by producing a rich baseline and historical coverage of knowledge about the compound. However, as a whole, this approach does not necessarily ensure an accurate reflection of the most current research being performed in the toxicology realm at large. In 2011, CTD collaborated with Pfizer, Inc., to curate an additional ∼80 000 articles selected by Pfizer for information regarding interactions between therapeutic compounds and four disease subsets (cardiovascular, neurological, renal and hepatic defects); of these papers, 53 951 contained curatable data. This Pfizer-driven curation is now freely available to all users and integrated with core CTD. Both of these initial corpora (core CTD and the Pfizer set) focused on achieving data completeness for specific types of information by retrieving articles regardless of when the data were published.
Keeping CTD current for its users with the most recent scientific results is a challenging process, especially with respect to the encroaching ‘data deluge’ (7). Towards that end, CTD has successfully implemented numerous processes to make manual curation as efficient and productive as possible, including a rigorous training period for new biocurators (5), measuring baseline curation metrics (8), incorporating practical controlled vocabularies as part of our curation paradigm (9), developing a highly efficient web-based curation tool for our remote biocurators (5) and successfully implementing text-mining tools to help prioritize and rank the most relevant articles for curation (8). To address the issue of data currency, we report here a pilot project of curating targeted journals based on publication date (as opposed to information content) as a means to more accurately reflect the current research interests in the toxicological community as a whole. Targeted journal curation should complement our chemical-centric approach to help balance and advance both data currency and completeness at CTD.
Data Currency
At CTD, data currency can be approximated by establishing the ‘age’ of data, reflected by the publication year of the article from whence the information was extracted. At the start of this experiment (March 2012), CTD included curated content from 88 035 articles published over the last 66 years, from 1946 to 2012. The manual curation paradigm for CTD was developed and implemented starting in 2005; consequently, we refer to articles with publication dates before 2003 as ‘legacy literature’, articles published within the last 2 full years (here, 2010–2012) as ‘current literature’ and articles published in the intervening time as ‘contemporary literature’. Using these arbitrary ranges, the age of data for CTD can be described as 46 113 (52%) legacy articles, 36 900 (42%) contemporary articles and 5022 (6%) current articles (Figure 1).
![Data currency at CTD. In March 2012, CTD contained 88 035 articles published between 1946 and 2012, including 46 113 (52%) legacy articles (grey), 36 900 (42%) contemporary articles (blue) and 5022 (6%) current articles (red); for simplicity, the number of articles for publication years 1946–66 were condensed into a single bar. When the number of curated articles in CTD is compared against an approximate number of available toxicogenomic articles from PubMed (solid black line), a noticeable hypothetical minimum gap in data currency is seen, especially for years 2010–11 (dashed lines). To approximate the number of hypothetical toxicogenomic articles for each year, PubMed was queried with the generic string: (toxicology OR toxicogenomics) OR [chemical AND (gene OR mRNA OR transcript)] NOT review[pt] AND ("YYYY/01/01"[PPDAT]:"YYYY/12/31"[PPDAT]), where YYYY = year of interest. The retrieved background is clearly an underrepresentation of the possible available literature (perhaps by as much as 2-fold; (1)); thus, the gap in data currency is a described as a ‘minimum gap’.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/database/2012/10.1093_database_bas051/1/m_bas051f1p.jpeg?Expires=1773786858&Signature=DHvDQCtsB9tynmkR7HYiYIuvRy6tQWbs1sZqJTYbgoUrt9yN40z9vPGOQLhLZYA75InocDeYHVLPOnpU3D9-MB0SjTmtk2~xxBzBf3QqrXQCwrcoCjlxBul9PZi6qSWshM7p4w-k1JNWsazkLkGD5pgH8ZlCZs7DY1mRIhjb2K0XT3SooA941P~w9ZpTvmzB1af8qmaZpezwcaEA7H8QeZBLqJnBckpLz31fqXoC6Z8~5vud~KyZyTmUv-syhbJzS9wRkywH1SEQKHUAqO8S~D78v18Dch50CQfGwfF55-mL2z41ifcVtLOj8SENgw3QSFnGkpUAgk4lPpMOk6nuRw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
Data currency at CTD. In March 2012, CTD contained 88 035 articles published between 1946 and 2012, including 46 113 (52%) legacy articles (grey), 36 900 (42%) contemporary articles (blue) and 5022 (6%) current articles (red); for simplicity, the number of articles for publication years 1946–66 were condensed into a single bar. When the number of curated articles in CTD is compared against an approximate number of available toxicogenomic articles from PubMed (solid black line), a noticeable hypothetical minimum gap in data currency is seen, especially for years 2010–11 (dashed lines). To approximate the number of hypothetical toxicogenomic articles for each year, PubMed was queried with the generic string: (toxicology OR toxicogenomics) OR [chemical AND (gene OR mRNA OR transcript)] NOT review[pt] AND ("YYYY/01/01"[PPDAT]:"YYYY/12/31"[PPDAT]), where YYYY = year of interest. The retrieved background is clearly an underrepresentation of the possible available literature (perhaps by as much as 2-fold; (1)); thus, the gap in data currency is a described as a ‘minimum gap’.
Although current literature will typically be underrepresented as a percentage of a database as a whole due to the ephemeral nature of what is considered current, it is important for CTD to report the latest scientific results to our users in a timely manner. To estimate the data currency gap in CTD, we interrogated PubMed (http://www.ncbi.nlm.nih.gov/pubmed) with a broad toxicogenomics query string, restricted by each year for 2003–11, to estimate the number of possible toxicogenomic papers available in PubMed. Since it is difficult to derive a query that could retrieve a complete corpus of articles containing all toxicogenomic data, these results should be considered an underrepresentation. Nonetheless, the query helps approximate a minimum background estimate as to the number of potentially curatable articles (Figure 1). By comparing the number of curated articles in CTD against this hypothetical background, we can estimate a minimum gap in data currency at CTD (Figure 1). While not completely accurate, this method nonetheless clearly shows a noticeably increasing gap discrepancy in the current literature in CTD versus the hypothetical toxicology literature available from PubMed, especially for the years 2010–11.
To improve data currency at CTD, we decided to test the feasibility of ‘target journal curation’, wherein curators would be assigned to review and curate selected, current journals from cover-to-cover each month, as a means to accurately report the current state of toxicology research as a whole. This approach helps to skirt any ‘chemical bias’ due to a chemical-centric approach and should represent a better snapshot of the information being produced in the toxicology community while improving data currency in CTD.
Targeted Journal Curation
CTD has actually been using targeted journal curation since May 2007 on a small scale with the journal Nature Genetics (http://www.nature.com/ng/), as a means to keep up-to-date with the most current gene–disease interactions. Nature Genetics is an ideal journal for this practice, since it publishes each month’s issue online prior to the start of the month, and by the time of that publication, each article has been issued a PubMed identification number (PMID), a mandatory identifier in CTD. This timely and ordered process allows Nature Genetics articles to be readily retrieved from the web and manually curated into CTD on a month-by-month basis with no delay. To test the feasibility of targeted journal curation on a larger scale and beyond the scope of just disease information, CTD needed first to select journals that would be most relevant to its toxicogenomic mission.
Journal selection
In June 2009, we ranked the top 127 journals represented in CTD using three criteria: the overall number of curated articles, the number of annotated interactions and an index score based on the averaged number of interactions extracted per article. From this initial list, we selected three journals for this pilot project (Table 1). The top two journals with the highest index score were Toxicological Sciences (TS; http://toxsci.oxfordjournals.org/), the official journal of the Society of Toxicology, and Environmental Health Perspectives (EHP; http://ehp03.niehs.nih.gov/). We also selected Chemico-Biological Interactions (CBI; http://www.journals.elsevier.com/chemico-biological-interactions), a ‘mid-level’ journal with respect to our ranking criteria. Although CBI has both a lower impact factor and CTD ranking compared with TS and EHP (Table 1), it has been publishing since 1969 and places an emphasis on research that elucidates molecular mechanisms of toxicology for characterized chemicals, a scope that accurately parallels CTD’s curation paradigm and mission (3).
| Journal . | Publisher . | Available online archive . | CTD rank, for number of curated articlesa . | CTD rank, for number of curated interactionsa . | CTD rank, for index scorea . | Impact factorb . | Scopeb . |
|---|---|---|---|---|---|---|---|
| Toxicological Sciences (TS) | Oxford University Press | 1981 to current | #3 | #1 | #1 | 5.09 | ‘All areas of toxicology’ |
| Environmental Health Perspectives (EHP) | National Institute of Environmental Health Sciences | 1972 to current | #25 | #7 | #2 | 6.09 | ‘Interrelationships between the environment and human health’ |
| Chemico-Biological Interactions (CBI) | Elsevier Ireland Ltd | 1969 to current | #32 | #49 | #50 | 2.83 | ‘Toxicological mechanisms associated with interactions between chemicals and biological systems’ |
| Journal . | Publisher . | Available online archive . | CTD rank, for number of curated articlesa . | CTD rank, for number of curated interactionsa . | CTD rank, for index scorea . | Impact factorb . | Scopeb . |
|---|---|---|---|---|---|---|---|
| Toxicological Sciences (TS) | Oxford University Press | 1981 to current | #3 | #1 | #1 | 5.09 | ‘All areas of toxicology’ |
| Environmental Health Perspectives (EHP) | National Institute of Environmental Health Sciences | 1972 to current | #25 | #7 | #2 | 6.09 | ‘Interrelationships between the environment and human health’ |
| Chemico-Biological Interactions (CBI) | Elsevier Ireland Ltd | 1969 to current | #32 | #49 | #50 | 2.83 | ‘Toxicological mechanisms associated with interactions between chemicals and biological systems’ |
aAs of June 2009 (range #1–127 with #1 being the highest). bSelf-reported on journal’s website (April 2012).
| Journal . | Publisher . | Available online archive . | CTD rank, for number of curated articlesa . | CTD rank, for number of curated interactionsa . | CTD rank, for index scorea . | Impact factorb . | Scopeb . |
|---|---|---|---|---|---|---|---|
| Toxicological Sciences (TS) | Oxford University Press | 1981 to current | #3 | #1 | #1 | 5.09 | ‘All areas of toxicology’ |
| Environmental Health Perspectives (EHP) | National Institute of Environmental Health Sciences | 1972 to current | #25 | #7 | #2 | 6.09 | ‘Interrelationships between the environment and human health’ |
| Chemico-Biological Interactions (CBI) | Elsevier Ireland Ltd | 1969 to current | #32 | #49 | #50 | 2.83 | ‘Toxicological mechanisms associated with interactions between chemicals and biological systems’ |
| Journal . | Publisher . | Available online archive . | CTD rank, for number of curated articlesa . | CTD rank, for number of curated interactionsa . | CTD rank, for index scorea . | Impact factorb . | Scopeb . |
|---|---|---|---|---|---|---|---|
| Toxicological Sciences (TS) | Oxford University Press | 1981 to current | #3 | #1 | #1 | 5.09 | ‘All areas of toxicology’ |
| Environmental Health Perspectives (EHP) | National Institute of Environmental Health Sciences | 1972 to current | #25 | #7 | #2 | 6.09 | ‘Interrelationships between the environment and human health’ |
| Chemico-Biological Interactions (CBI) | Elsevier Ireland Ltd | 1969 to current | #32 | #49 | #50 | 2.83 | ‘Toxicological mechanisms associated with interactions between chemicals and biological systems’ |
aAs of June 2009 (range #1–127 with #1 being the highest). bSelf-reported on journal’s website (April 2012).
Article selection and delivery
Three years of articles (publication dates from 2009–11) for TS, EHP and CBI were collected for manual curation, resulting in a corpus of 2425 articles (Table 2). Journal articles were pre-selected by removing documents that CTD does not routinely curate, such as review articles, letters to editors, highlights and essays, to help concentrate specifically on original, peer-reviewed research reports. Two different methods of article delivery to the biocurators were tested.
| Metric . | TS (2009–11) . | CBI (2009–11) . | EHP (2009–11) . | Total . | Averagef . |
|---|---|---|---|---|---|
| No. articles examined | 884 | 785 | 756 | 2425 | n/a |
| No. articles curated by biocurator | 493 | 485 | 274 | 1252 | n/a |
| No. articles found to have been previously curated | 107 | 88 | 30 | 225 | n/a |
| No. articles rejected | 284 | 212 | 452 | 948 | n/a |
| % Curated articlesa | 68% | 73% | 40% | n/a | 61% |
| No. chemical–gene interactions curatedb | 40 992 | 5176 | 4468 | 50 636 | n/a |
| No. chemical–disease and gene–disease interactions curatedb | 835 | 839 | 442 | 2116 | n/a |
| Total no. interactions curatedb | 41 827 | 6015 | 4910 | 52 752 | n/a |
| Time spent on curatable articles, adjusted (min)c | 25 405 | 11 950 | 5199 | 42 554 | n/a |
| Curation rate, adjusted (minutes per curatable article)d | 51.5 | 24.6 | 19.0 | n/a | 34.0 |
| Interaction yield rate (interactions per minute)e | 1.6 | 0.5 | 0.9 | n/a | 1.2 |
| Metric . | TS (2009–11) . | CBI (2009–11) . | EHP (2009–11) . | Total . | Averagef . |
|---|---|---|---|---|---|
| No. articles examined | 884 | 785 | 756 | 2425 | n/a |
| No. articles curated by biocurator | 493 | 485 | 274 | 1252 | n/a |
| No. articles found to have been previously curated | 107 | 88 | 30 | 225 | n/a |
| No. articles rejected | 284 | 212 | 452 | 948 | n/a |
| % Curated articlesa | 68% | 73% | 40% | n/a | 61% |
| No. chemical–gene interactions curatedb | 40 992 | 5176 | 4468 | 50 636 | n/a |
| No. chemical–disease and gene–disease interactions curatedb | 835 | 839 | 442 | 2116 | n/a |
| Total no. interactions curatedb | 41 827 | 6015 | 4910 | 52 752 | n/a |
| Time spent on curatable articles, adjusted (min)c | 25 405 | 11 950 | 5199 | 42 554 | n/a |
| Curation rate, adjusted (minutes per curatable article)d | 51.5 | 24.6 | 19.0 | n/a | 34.0 |
| Interaction yield rate (interactions per minute)e | 1.6 | 0.5 | 0.9 | n/a | 1.2 |
aIncludes articles curated and previously curated. bDoes not include data from articles previously curated. cAdjusted time removes estimated minutes spent on rejected articles, which averages 2.5 min per rejected article [see (8)]. dAdjusted curation rate = adjusted curation time divided by no. articles curated by biocurator. eInteraction yield rate = total no. interactions divided by adjusted curation time. fMacro-averages derived from values in Total column; n/a = not applicable.
| Metric . | TS (2009–11) . | CBI (2009–11) . | EHP (2009–11) . | Total . | Averagef . |
|---|---|---|---|---|---|
| No. articles examined | 884 | 785 | 756 | 2425 | n/a |
| No. articles curated by biocurator | 493 | 485 | 274 | 1252 | n/a |
| No. articles found to have been previously curated | 107 | 88 | 30 | 225 | n/a |
| No. articles rejected | 284 | 212 | 452 | 948 | n/a |
| % Curated articlesa | 68% | 73% | 40% | n/a | 61% |
| No. chemical–gene interactions curatedb | 40 992 | 5176 | 4468 | 50 636 | n/a |
| No. chemical–disease and gene–disease interactions curatedb | 835 | 839 | 442 | 2116 | n/a |
| Total no. interactions curatedb | 41 827 | 6015 | 4910 | 52 752 | n/a |
| Time spent on curatable articles, adjusted (min)c | 25 405 | 11 950 | 5199 | 42 554 | n/a |
| Curation rate, adjusted (minutes per curatable article)d | 51.5 | 24.6 | 19.0 | n/a | 34.0 |
| Interaction yield rate (interactions per minute)e | 1.6 | 0.5 | 0.9 | n/a | 1.2 |
| Metric . | TS (2009–11) . | CBI (2009–11) . | EHP (2009–11) . | Total . | Averagef . |
|---|---|---|---|---|---|
| No. articles examined | 884 | 785 | 756 | 2425 | n/a |
| No. articles curated by biocurator | 493 | 485 | 274 | 1252 | n/a |
| No. articles found to have been previously curated | 107 | 88 | 30 | 225 | n/a |
| No. articles rejected | 284 | 212 | 452 | 948 | n/a |
| % Curated articlesa | 68% | 73% | 40% | n/a | 61% |
| No. chemical–gene interactions curatedb | 40 992 | 5176 | 4468 | 50 636 | n/a |
| No. chemical–disease and gene–disease interactions curatedb | 835 | 839 | 442 | 2116 | n/a |
| Total no. interactions curatedb | 41 827 | 6015 | 4910 | 52 752 | n/a |
| Time spent on curatable articles, adjusted (min)c | 25 405 | 11 950 | 5199 | 42 554 | n/a |
| Curation rate, adjusted (minutes per curatable article)d | 51.5 | 24.6 | 19.0 | n/a | 34.0 |
| Interaction yield rate (interactions per minute)e | 1.6 | 0.5 | 0.9 | n/a | 1.2 |
aIncludes articles curated and previously curated. bDoes not include data from articles previously curated. cAdjusted time removes estimated minutes spent on rejected articles, which averages 2.5 min per rejected article [see (8)]. dAdjusted curation rate = adjusted curation time divided by no. articles curated by biocurator. eInteraction yield rate = total no. interactions divided by adjusted curation time. fMacro-averages derived from values in Total column; n/a = not applicable.
Method 1: Journal Online Archive (TS)
For TS, biocurators used the journal’s archived web portal to curate their assigned year (http://toxsci.oxfordjournals.org/content/by/year). For example, the biocurator would select the first issue of January 2009 and work through the table of contents, curating each article in a progressive manner. The biocurator first reviewed the online abstract to make the decision of whether or not the article was ‘curatable’ for CTD criteria (5). If the article warranted curation, the biocurator searched PubMed using the article’s title to retrieve its PMID and begin curation with CTD’s Curation Tool (5). While the number of articles rejected as ‘non-curatable’ was recorded, the specific PMIDs of those rejected articles were not queried for nor saved. Once the biocurator finished with 1 month, they would close that issue and open up the next month’s to repeat the process.
Method 2: PubMed Queries (CBI and EHP)
For CBI and EHP journals, a project manager first queried PubMed to download a file of PMIDs for an entire year of research articles using PubMed codes for specific journal titles (code = [JOUR]), publication years (code = [PPDAT]), article types (code = [PT]) and presence of abstracts (code = has abstract[TEXT]) (10). This file of PMIDs was then sent to the biocurator who used the PubMed interface to retrieve the abstracts to begin curation, obviating the need to use the journal’s own web portal or to find the PMID on their own.
In both methods, biocurators recorded the number of articles that ended up being curatable; the number of articles found to have been previously curated and the number of rejected articles that did not contain chemical–gene, chemical–disease or gene–disease interactions. Overall, Method 2 was determined to be more advantageous because it eliminated the need for the biocurator to first find the PMID on their own, it allowed the PMID of rejected articles to be recorded for future insight into training sets for text-mining opportunities, and it allowed biocurators to work more efficiently using just one file of PMIDs instead of having to open up new monthly issues from the journal’s online archive.
Curation metrics
To be curatable for CTD, an article must describe a chemical–gene, chemical–disease or gene–disease interaction, where we use the word ‘gene’ to refer to any gene aspect, including mRNA, protein, promoter, exon, untranslated region, etc. (3). CTD biocurators read and curate the significant main points emphasized by the authors in the abstract. However, it is often necessary for the biocurator to go to the full text in order to resolve ambiguities found in the abstract, such as the correct species or gene identity. Once in the full text, the biocurator may capture important additional data not found in the abstract, including relevant information from the Supplementary tables (e.g. microarray tables). Thus, while CTD biocurators try to curate exclusively from the abstract whenever possible, they are not restricted to only the abstract; when necessary, the biocurator is allowed to go to the full text to resolve ambiguities and curate additional, significant data alluded to in the abstract. While entering interactions in the online Curation Tool, biocurators also designate the source of the interaction as either being derived from the ‘abstract’ or the ‘full text’ (5).
The combined corpus from 2009–11 for all three targeted journals was 2425 articles. During review, 225 of these articles had been previously curated for CTD and 948 articles were rejected as non-curatable, leaving 1252 curatable articles from which 52 752 total interactions were manually extracted (Table 2). Of these 52 752 interactions, 49 552 of them (94%) were novel interactions not yet represented in CTD. This substantial addition of new content supports the value in maintaining data currency. By analysing each journal independently, interesting patterns are seen. For example, TS, CBI and EHP averaged a total of 68, 73 and 40% curatable articles, respectively, indicating that TS and CBI each provided significantly more curatable papers than did EHP. Also, the averaged times to curate an article differed between journals: 51.5 (TS), 24.6 (CBI) and 19.0 (EHP) min per article (Table 2). Overall, TS articles were more time-consuming to curate primarily due to the presence of many papers describing microarray technology to assay the effects of chemical exposure on gene expression. These articles provide relatively numerous chemical–gene interactions to capture, as reflected by the dramatically higher number of chemical–gene interactions curated from all three issues of TS (40 992) compared with CBI (5176) and EHP (4468). Although the CTD Curation Tool accommodates the use of spreadsheets to enable biocurators to upload high-volume data (such as microarray results) in an efficient fashion (5), it nevertheless takes a much longer time to curate these microarray articles. Thus, to get a better perspective of efficiency and productivity, we calculated for each targeted journal an ‘interaction yield rate’, which describes the number of interactions manually curated per unit of time (Table 2). Here, TS articles averaged an interaction yield rate of 1.6 interactions per minute, compared with 0.5 and 0.9 interactions per minute for CBI and EHP, respectively, demonstrating that TS articles provide ∼2-fold greater yield of data for the same amount of time invested.
Data currency improvement
For publication year 2009, targeted journal curation provided new data from 345 articles (112 from TS, 134 from CBI and 99 from EHP; Table 3). These 345 articles help close the hypothetical minimum gap in data currency (2053 gap articles; Figure 1) for that year by 17%. Likewise, for publication year 2010, targeted journal curation added 498 combined new articles from all three journals, reducing that year’s hypothetical data currency gap by 16%. Finally, for 2011, targeted journal curation helped improve the data currency gap by 7% via the addition of curated content from 409 new articles into CTD. However, since the hypothetical toxicology background may be off by at least 2-fold ( A.P. Davis, unpublished data), we can alternatively measure improvement in data currency by considering the number of new articles added to that year’s number of publications currently in CTD. Thus, targeted journal curation added 345 new articles to the 4332 articles currently in CTD for the year 2009 (Figure 1), representing an 8% increase. For 2010, the 498 added articles produced a 10% increase for that year, and for 2011, the 409 new articles resulted in a 27% increase. Considering that this pilot project was conducted by just three CTD biocurators in only 7 weeks, we should be able to rapidly and significantly increase the data currency at CTD by expanding targeted journal curation to include more CTD biocurators and additional journals (see ‘Discussion’ section).
| Journal (year) . | No. newly curated articlesa . | No. previously curated articlesb . | Total no. curated articlesc . | No. chemicalsc . | No. genesc . | No. diseasesc . |
|---|---|---|---|---|---|---|
| TS (2009) | 112 | 57 | 169 | 276 | 7776 | 80 |
| TS (2010) | 187 | 37 | 224 | 443 | 10 407 | 102 |
| TS (20,11) | 194 | 13 | 207 | 467 | 8760 | 129 |
| CBI (2009) | 134 | 65 | 199 | 429 | 433 | 67 |
| CBI (2010) | 230 | 17 | 247 | 480 | 705 | 128 |
| CBI (2011) | 121 | 6 | 127 | 269 | 215 | 37 |
| EHP (2009) | 99 | 9 | 108 | 141 | 1889 | 80 |
| EHP (2010) | 81 | 12 | 93 | 110 | 413 | 53 |
| EHP (2011) | 94 | 9 | 103 | 232 | 1282 | 61 |
| Journal (year) . | No. newly curated articlesa . | No. previously curated articlesb . | Total no. curated articlesc . | No. chemicalsc . | No. genesc . | No. diseasesc . |
|---|---|---|---|---|---|---|
| TS (2009) | 112 | 57 | 169 | 276 | 7776 | 80 |
| TS (2010) | 187 | 37 | 224 | 443 | 10 407 | 102 |
| TS (20,11) | 194 | 13 | 207 | 467 | 8760 | 129 |
| CBI (2009) | 134 | 65 | 199 | 429 | 433 | 67 |
| CBI (2010) | 230 | 17 | 247 | 480 | 705 | 128 |
| CBI (2011) | 121 | 6 | 127 | 269 | 215 | 37 |
| EHP (2009) | 99 | 9 | 108 | 141 | 1889 | 80 |
| EHP (2010) | 81 | 12 | 93 | 110 | 413 | 53 |
| EHP (2011) | 94 | 9 | 103 | 232 | 1282 | 61 |
aFrom targeted journal curation. bPreviously curated in CTD before targeted journal curation. cProvided in the Supplementary Data.
| Journal (year) . | No. newly curated articlesa . | No. previously curated articlesb . | Total no. curated articlesc . | No. chemicalsc . | No. genesc . | No. diseasesc . |
|---|---|---|---|---|---|---|
| TS (2009) | 112 | 57 | 169 | 276 | 7776 | 80 |
| TS (2010) | 187 | 37 | 224 | 443 | 10 407 | 102 |
| TS (20,11) | 194 | 13 | 207 | 467 | 8760 | 129 |
| CBI (2009) | 134 | 65 | 199 | 429 | 433 | 67 |
| CBI (2010) | 230 | 17 | 247 | 480 | 705 | 128 |
| CBI (2011) | 121 | 6 | 127 | 269 | 215 | 37 |
| EHP (2009) | 99 | 9 | 108 | 141 | 1889 | 80 |
| EHP (2010) | 81 | 12 | 93 | 110 | 413 | 53 |
| EHP (2011) | 94 | 9 | 103 | 232 | 1282 | 61 |
| Journal (year) . | No. newly curated articlesa . | No. previously curated articlesb . | Total no. curated articlesc . | No. chemicalsc . | No. genesc . | No. diseasesc . |
|---|---|---|---|---|---|---|
| TS (2009) | 112 | 57 | 169 | 276 | 7776 | 80 |
| TS (2010) | 187 | 37 | 224 | 443 | 10 407 | 102 |
| TS (20,11) | 194 | 13 | 207 | 467 | 8760 | 129 |
| CBI (2009) | 134 | 65 | 199 | 429 | 433 | 67 |
| CBI (2010) | 230 | 17 | 247 | 480 | 705 | 128 |
| CBI (2011) | 121 | 6 | 127 | 269 | 215 | 37 |
| EHP (2009) | 99 | 9 | 108 | 141 | 1889 | 80 |
| EHP (2010) | 81 | 12 | 93 | 110 | 413 | 53 |
| EHP (2011) | 94 | 9 | 103 | 232 | 1282 | 61 |
aFrom targeted journal curation. bPreviously curated in CTD before targeted journal curation. cProvided in the Supplementary Data.
Intra-journal comparison and research sub-specialties
For all subsequent journal analysis (both intra-journal and inter-journal), we first combined data from the 1252 curated articles described above with data from the 225 articles that had been previously curated from these three test journals in CTD at an earlier time. This combined data more accurately represents the entire knowledge space for the targeted journals during the 2009–11 publication range. The number of chemicals, genes and diseases were identified for each journal set (Table 3 and see the Supplementary Data).
We used CTD’s ‘MyVenn’ diagram analytical tool (http://ctdbase.org/tools/myVenn.go) to perform an intra-journal comparison of the chemicals, genes and diseases reported for the three different publication years 2009–11 for all three targeted journals (Figure 2). One goal of a journal-centric approach to curation is to help avoid the inherent chemical bias that results from an exclusive chemical-centric approach. However, the act of targeting selected journals itself also biases curation, since different journals publish in different sub-specialties of toxicology.
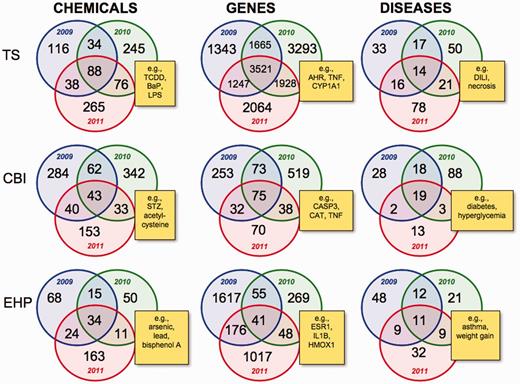
Intra-journal data comparison for 2009–11. Nine Venn diagrams depict the overlapping datasets for the number of chemicals, genes and diseases for each journal for publication years 2009 (blue circles), 2010 (green circles) and 2011 (red circles). Yellow boxes provide examples of shared elements for all 3 years in the centre intersection of each Venn diagram and are described in the main text. TS = Toxicological Sciences, CBI = Chemico-Biological Interactions and EHP = Environmental Health Perspectives. All data are provided in the Supplementary Data, and readers can use CTD’s ‘MyVenn’ tool (http://ctdbase.org/tools/myVenn.go) to re-draw the Venn diagrams to explore all the sets.
For example, curated data from all 3 years from TS articles show an overlap of 88 chemicals (Figure 2), including tetrachlorodibenzodioxin (TCDD), lipopolysaccharides (LPS), acetaminophen and benzo(a)pyrene (BaP). In the disease comparisons, there are 14 commonly curated disorders from 2009–11 including drug-induced liver injury (DILI), inflammation, necrosis and neurotoxicity syndromes, and the most commonly curated genes from TS articles include AHR, CYP1A1, TNF and CASP3 (Figure 2). A common research theme for TS articles is seen with known network connections involving TCDD-AHR-CYP1A1, BaP-AHR-CYP1A1, acetaminophen-DILI, LPS-TNF-inflammation and CASP3-necrosis.
For CBI, some of the shared 43 chemicals reported from 2009–11 include plant extracts, streptozocin (STZ), and acetylcysteine, while the overlapping 19 diseases consist of experimentally induced diabetes (which can be chemically induced by STZ) and hyperglycemia (Figure 2). CASP3, CAT and TNF are three of the most commonly curated genes from CBI.
The journal EHP, on the other hand, clearly reveals a distinct sub-specialty, with its 34 overlapping chemicals focusing on more commonly encountered environmental compounds such as particulate matter, bisphenol A, arsenic and lead (Figure 2). The shared 11 diseases also reflect better known environmental disorders, including prenatal exposure delayed effects, weight gain and asthma.
Inter-journal comparison and areas of environmental health
Next, we performed an inter-journal comparison for each publication year to look for common elements to all three journals from 2009 to 2011. From 2009 to 2011, there are 16, 24 and 14 chemicals, respectively, shared by all three targeted journals (Figure 3). Similarly, there are 95, 85 and 76 common genes for years 2009, 2010 and 2011 from these journals, respectively (Figure 4), and 4, 11 and 6 common diseases, respectively (Figure 5). We compared these overlapping sets for each year to look for prominent trends in environmental health research.
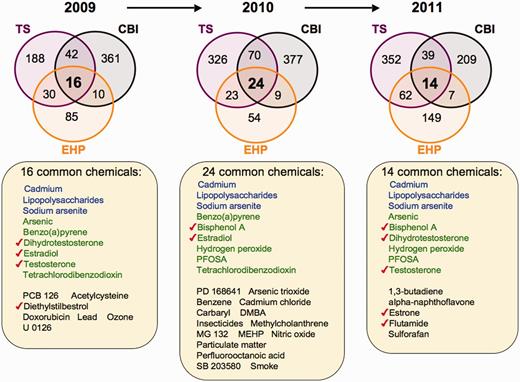
Prominent environmental chemicals from inter-journal comparison. Three Venn diagrams depict the overlapping datasets for curated chemicals shared by journals TS (purple circles), CBI (black circles) and EHP (orange circles) for years 2009–11. The first three chemicals in each list (blue) are shared by all three journals for all 3 years, and nine chemicals (green) are shared in 2 of the 3 years. The other listed chemicals (black) are shared by the three journals for that unique year. Seven chemicals (red checks) are known to modulate sex hormone receptor signalling pathways. All data are provided in the Supplementary Data, and readers can use CTD’s ‘MyVenn’ tool (http://ctdbase.org/tools/myVenn.go) to re-draw the Venn diagrams to explore all the sets.
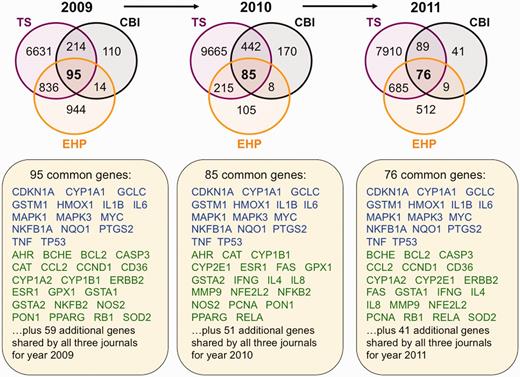
Trending toxicology gene sets from inter-journal comparison. Three Venn diagrams depict the overlapping datasets for curated genes shared by journals TS (purple circles), CBI (black circles) and EHP (orange circles) for years 2009–11. Fifteen genes (blue) are shared by all three journals for all 3 years, and 30 other genes (green) are shared in 2 of the 3 years. The additional genes specific for each individual year are not shown but listed as 59 (for 2009), 51 (for 2010) and 41 (for 2011). All data are provided in the Supplementary Data, and readers can use CTD’s ‘MyVenn’ tool (http://ctdbase.org/tools/myVenn.go) to re-draw the Venn diagrams to explore all the sets.
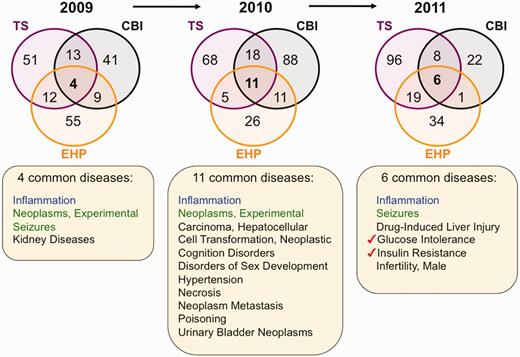
Environmental diseases from inter-journal comparison. Three Venn diagrams depict the overlapping datasets for curated diseases shared by journals TS (purple circles), CBI (black circles) and EHP (orange circles) for years 2009–11. Inflammation (blue) is shared by all three journals for all 3 years, and experimental neoplasms and seizures (green) are shared in 2 of the 3 years. The other listed diseases (black) are shared by the three journals for that unique year. In 2011, two pre-diabetes markers (red checks) are shared among all three journals. All data are provided in the Supplementary Data, and readers can use CTD’s ‘MyVenn’ tool (http://ctdbase.org/tools/myVenn.go) to re-draw the Venn diagrams to explore all the sets.
Figure 3 shows the compounds independently reported by all three journals for 2009–11. Three chemicals (cadmium, LPS and sodium arsenite) are common to all 3 years from all three journals. Nine other compounds are shared by 2 of the 3 years. Interestingly, there is an abundance of sex hormones distributed among all 3 years, including dihydrotestosterone, estradiol, testosterone, diethylstilbestrol and oestrone as well as two chemicals known to modulate sex hormone receptor signalling (bisphenol A and flutamide), supporting the increasing interest in hormone signalling in the toxicology community.
For 2009, there were 95 genes shared among all three journals (representing 1% of the TS gene set, 22% of the CBI set and 5% of the EHP set for that year). For 2010, there were 85 genes shared among all three journals (representing 0.8% of the TS gene set, 12% of the CBI set and 21% of EHP set for that year). For 2011, there were 76 genes shared among all three journals (representing 0.9% of the TS gene set, 35% of the CBI set and 6% of EHP set). Of these common genes, there are 15 genes shared by all three journals for all 3 years, and 30 other genes shared by 2 of the 3 years (Figure 4). Together, these 45 genes might represent trending toxicological genes of interest; alternatively, they might just simply be genes that are commonly studied (or easily assayed). We analysed these genes using CTD’s ‘Gene Set Enricher’ tool (http://ctdbase.org/tools/enricher.go), which finds the enriched Gene Ontology (GO) or Pathway terms for a gene list. The top 10 GO biological process terms enriched for the 45 genes include seven terms describing a stimulus response, such as ‘response to chemical stimulus’ (GO:0042221), ‘response to drug’ (GO:0042493) and ‘response to stress’ (GO:0006950), supporting the idea that these genes are of toxicological value (Table 4).
| GO terma . | Corrected P-value b . |
|---|---|
| Response to chemical stimulus | 2.07E−42 |
| Cellular response to chemical stimulus | 1.22E−36 |
| Response to organic substance | 1.09E−32 |
| Cellular response to stimulus | 2.19E−31 |
| Response to stimulus | 2.96E−29 |
| Response to drug | 2.02E−27 |
| Response to stress | 1.81E−26 |
| Positive regulation of biological process | 2.37E−26 |
| Regulation of cell proliferation | 3.45E−26 |
| Cell proliferation | 3.85E−26 |
| GO terma . | Corrected P-value b . |
|---|---|
| Response to chemical stimulus | 2.07E−42 |
| Cellular response to chemical stimulus | 1.22E−36 |
| Response to organic substance | 1.09E−32 |
| Cellular response to stimulus | 2.19E−31 |
| Response to stimulus | 2.96E−29 |
| Response to drug | 2.02E−27 |
| Response to stress | 1.81E−26 |
| Positive regulation of biological process | 2.37E−26 |
| Regulation of cell proliferation | 3.45E−26 |
| Cell proliferation | 3.85E−26 |
aRetrieved 8 May 2012 (CTD version 11 146). bBonferroni multiple testing adjustment.
| GO terma . | Corrected P-value b . |
|---|---|
| Response to chemical stimulus | 2.07E−42 |
| Cellular response to chemical stimulus | 1.22E−36 |
| Response to organic substance | 1.09E−32 |
| Cellular response to stimulus | 2.19E−31 |
| Response to stimulus | 2.96E−29 |
| Response to drug | 2.02E−27 |
| Response to stress | 1.81E−26 |
| Positive regulation of biological process | 2.37E−26 |
| Regulation of cell proliferation | 3.45E−26 |
| Cell proliferation | 3.85E−26 |
| GO terma . | Corrected P-value b . |
|---|---|
| Response to chemical stimulus | 2.07E−42 |
| Cellular response to chemical stimulus | 1.22E−36 |
| Response to organic substance | 1.09E−32 |
| Cellular response to stimulus | 2.19E−31 |
| Response to stimulus | 2.96E−29 |
| Response to drug | 2.02E−27 |
| Response to stress | 1.81E−26 |
| Positive regulation of biological process | 2.37E−26 |
| Regulation of cell proliferation | 3.45E−26 |
| Cell proliferation | 3.85E−26 |
aRetrieved 8 May 2012 (CTD version 11 146). bBonferroni multiple testing adjustment.
To look for prominent environmental diseases, we compared the shared sets of diseases for all three journals from 2009 to 2011 (Figure 5). Inflammation is the one common disease seen in TS, CBI and EHP for this time period, with experimental neoplasms and seizures being shared by 2 of the 3 years. In 2011, only six common diseases were distributed over the three journals; of those six, however, two of them (glucose intolerance and insulin resistance) are markers of pre-diabetes.
Discussion
Different databases select articles for curation via different techniques. Established databases tend to focus on the current literature (6,11,12) but also retro-curate the legacy literature to improve data completeness for certain categories, such as mutant phenotypes (13) or specific sets of genes (14). As a relatively new database, CTD has focused on data completeness for individual chemicals (using legacy literature) to help build a historic, solid knowledge foundation about interactions between chemicals, genes and diseases, in addition to maintaining data currency (using recent literature) to cover the topical interests of the toxicology research community. Whereas model organism databases can probe the current literature by running periodic PubMed queries for new papers citing their species of interest (6,14,15), CTD data are not isolated to a single species, but rather to all eumetazoans as well as any chemical or disease (16). This broad coverage makes it challenging for CTD to accurately design a single-generic query to interrogate the current literature on a routine basis.
In the past, CTD has used a Chemical Priority Matrix to select chemicals of interest to the toxicology community for prioritizing literature for manual curation. This approach, used since 2005, has produced a solid foundation of knowledge and data completeness for >800 chemicals. CTD curates data for ‘all’ chemicals encountered in any article, regardless of whether the compound was the triaged chemical-of-interest (5). This practice results in the added curation of numerous ‘secondary chemicals’. As of March 2012, CTD included data for 820 priority chemicals and partial data for 7481 secondary chemicals. However, even after a chemical-of-interest has undergone priority triaging for data completeness, that chemical will nonetheless become out-of-date with time, requiring re-curation at scheduled intervals to maintain data completeness for the chemical. Naturally, this chemical-centric approach leads to a ‘chemical bias’, wherein overall CTD knowledge is skewed towards specific types of compounds. To balance this bias, we found our targeted journal curation method is a more manageable solution to improve data currency.
In 7 weeks, three CTD biocurators manually reviewed 2425 articles from 3 years worth of three targeted journals, extracting 52 752 interactions from 1252 curatable articles and improving the overall data currency by closing a hypothetical gap by 7–17% for the years 2009–11 (bearing in mind that our hypothetical toxicology literature background may be off by 2-fold; A.P. Davis, unpublished data). Of the 52 752 interactions, 94% of them added novel content to CTD, highlighting the value and importance of maintaining data currency. The success of this pilot study encourages us to expand journal-centric curation as part of our regular curation pipeline (see below). By combining these complementary approaches into our workflow, we should improve both data completeness and data currency at CTD (Figure 6). As more journals are selected for inclusion, targeted journal curation should also help more evenly build content for chemicals, genes and diseases rather than biasing data solely towards chemicals-of-interest.
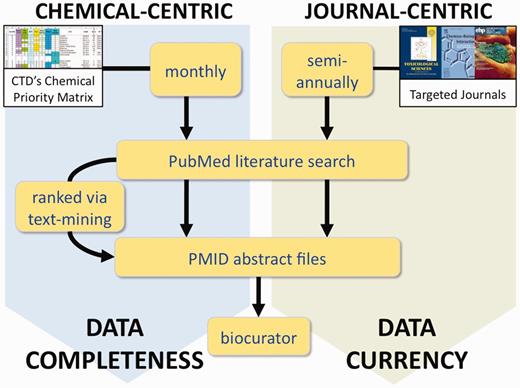
CTD’s two complementary processes for literature selection and curation. In the chemical-centric approach, each month we select several chemicals-of-interest from our Chemical Priority Matrix to query PubMed for all the literature (both current and legacy) for each chemical. Depending upon the size of the corpus, either all the abstracts are sent to the biocurator, or they are first processed through CTD’s text-mining algorithm to rank and prioritize the papers based upon data content. This approach results in data completeness for the chemical. In the journal-centric approach, we could retrieve the complete set of articles for selected targeted journals on a regular basis (perhaps semi-annually), providing a corpus of research papers that more accurately reflects the current state of toxicogenomics, regardless of any chemical bias. This method results in improved overall data currency at CTD.
Journal-centric curation will also facilitate data completeness and currency by finding new data for compounds that have already been curated at CTD via the chemical-centric method at an earlier time. For example, arsenic, one of CTD’s priority chemicals (4), has undergone three additional rounds of maintenance curation over the last 6 years. Implementing targeted journal curation, however, should help maintain both data completeness and currency for arsenic in a timelier manner by finding relevant articles as they are published, as opposed to waiting until the next scheduled maintenance.
Of the two methods tested for article delivery to the biocurator, we found that compiling a year’s worth of articles from a targeted journal into one file was more efficient and manageable than having biocurators individually access articles one-by-one from a journal’s website. Although this ‘one file’ approach is efficient, it does have the downside of delaying data currency. For example, a full year’s worth of 2011 journal articles could not be retrieved until at least January 2012. Performing the query at quarterly or monthly intervals can help ameliorate this delay, but would become difficult from a project management perspective, especially for a large number of journals (which often do not publish with the same periodicity). Querying on a semi-annual basis, however, may be a good compromise for balancing data currency with project management.
In addition to improving data currency and completeness for particular chemicals, targeted journal curation may also identify developing or prominent research trends. Common chemicals, genes and diseases from various journals for different years may dynamically reflect trending areas in environmental health. These results can be used to support or help develop and advance toxicology monitoring programs (17), public health and consumer awareness (18,19), and specialized microarrays to better study gene–environment interactions for personalized medicine (20). Here, in our limited analysis of only three journals from 2009 to 2011, we identified several chemicals of topical interest (cadmium, LPS, sodium arsenite and seven compounds known to activate sex hormone receptor signalling pathways), 45 toxicology genes (confirmed by enriched GO biological process annotations) and four common diseases (inflammation, experimental cancers, seizures and pre-diabetes). Expanding this meta-analysis to more journals and broader concepts may provide additional insight to developing research areas.
Selecting the most appropriate journals to target by CTD will be an iterative process. In June 2009, we analysed the top 127 journals represented in CTD and from that list we selected the three journals (TS, CBI and EHP) used in this pilot study. The results of our analysis confirm that different scientific journals often focus on specific research topics and publish different types of information. For example, the journal TS publishes many articles that use microarrays to assay how chemical exposure affects gene expression. With such papers, CTD biocurators only curate gene expression changes that the authors report as being statistically significant. Nonetheless, these articles are still time-consuming to curate, yet provide a higher interaction yield rate, suggesting TS may be a good candidate for increasing both data currency and data completeness efficiently. Curated interactions derived from high-throughput assays, such as microarrays, are additionally annotated by biocurators with an internal code (HTP). In a future release of CTD, users may have the option to filter interactions based upon their HTP status, in case some users prefer to exclude high-throughput data from their analysis. The journal EHP, on the other hand, had a low curatability index for core CTD data, with only 40% of the articles being curatable for chemical–gene–disease interactions; however, based upon feedback from the biocurators, this journal was found to be an excellent candidate for curating environmental exposure science, a new CTD initiative (21).
Going forward, CTD needs to identify additional journals beyond these three for targeted curation. A case-by-case analysis of individual journals may be too demanding. Instead, one practical solution may be to exploit the top 127 journals represented in CTD by simply searching for the word ‘tox’ in the journal title’s abbreviation, with the presumption that such journals would have a more devoted scope to toxicological research. We found that 19 of those 127 journals (15%) contain the string ‘tox’. As a proof of concept, we have since expanded journal-centric curation to include nine additional journals in our pipeline. Biocurators reviewed the most current articles from these nine journals while also working on other CTD projects. From March to August 2012, five biocurators added a total of 9631 articles to the database, of which 4254 (44%; including the 1252 articles described herein for the three journals TS, CBI and EHP, plus 3002 articles from the additional nine new journals) were derived exclusively from targeted journal curation. The remaining 5377 articles (56%) were curated for other CTD projects and had publication dates ranging from 1962 to 2012 (Figure 7). These results show that targeted journal curation can be easily and successfully incorporated into CTD’s pipeline without sacrificing the curation of legacy or contemporary articles for other projects. For year 2009, 77% of all newly added articles are derived exclusively from targeted journal curation (Figure 7). For 2010, it is 78%, for 2011, 80% and for 2012, it is 65%, representing a substantial increase in data currency. These nine new journals had publication years of 2009 up to the first half of 2012; going forward, however, as part of our regular pipeline, targeted journal curation would only have to focus on the most current year at hand and should be even faster to accommodate. As stated above, we envision targeted journal curation to be performed on a semi-annual basis, wherein a project manager will collect all the relevant PMIDs for all the targeted journals in 6-month intervals to allocate to the curation team.
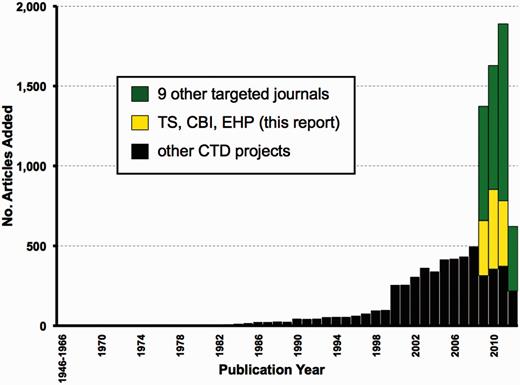
Expanding targeted journal curation at CTD. From March to August 2012, 9631 new articles were added to CTD. Of these, 4254 are from targeted journal curation, including 1252 from the three journals (TS, CBI and EHP) reported here (yellow bars) plus 3002 articles from nine additional journals (green bars) for publication years from 2009 to the first half of 2012. The remaining 5377 articles (black bars) are from other CTD projects and span publication years 1962–2012.
Undoubtedly, text-mining and machine-learning methods will also play an important role in literature selection (22,23). CTD has already successfully developed and implemented a text-mining algorithm that ranks selected articles with respect to relevant data content for any particular chemical-of-interest (8). As applied to current literature, text mining will help select the best articles for improving data currency, and when used to interrogate older literature, should also increase the efficiency of maintaining data completeness for particular chemicals.
Citing and Linking to CTD
To cite CTD, please see http://ctdbase.org/about/publications/#citing. Currently, over 28 external databases link to or present CTD data on their own websites. If you are interested in establishing links to CTD data, please notify us (http://ctdbase.org/help/contact.go) and follow these instructions: http://ctdbase.org/help/linking.jsp.
Funding
National Institute of Environmental Health Sciences (NIEHS): R01-ES014065 (Comparative Toxicogenomics Database) and R01-ES019604 (Generation of a centralized and integrated resource for exposure data). Funding for open access charge: NIEHS grants (R01-ES014065 and R01-ES019604).
Conflict of interest. None declared.
Acknowledgements
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
Author notes
Citation details: Allan Peter Davis, Robin J. Johnson, Kelley Lennon-Hopkins, Daniela Sciaky, Michael C. Rosenstein, Thomas C. Wiegers, and Carolyn J. Mattingly. Targeted journal curation as a method to improve data currency at the Comparative Toxicogenomics Database. Database (2012) Vol. 2012: article ID bas051; doi:10.1093/database/bas051.



