-
PDF
- Split View
-
Views
-
Cite
Cite
Amy L Olex, Evan French, Peter Burdette, Srilakshmi Sagiraju, Thomas Neumann, Tamas S Gal, Bridget T McInnes, TopEx: topic exploration of COVID-19 corpora - Results from the BioCreative VII Challenge Track 4, Database, Volume 2022, 2022, baac063, https://doi.org/10.1093/database/baac063
Close - Share Icon Share
Abstract
TopEx is a natural language processing application developed to facilitate the exploration of topics and key words in a set of texts through a user interface that requires no programming or natural language processing knowledge, thus enhancing the ability of nontechnical researchers to explore and analyze textual data. The underlying algorithm groups semantically similar sentences together followed by a topic analysis on each group to identify the key topics discussed in a collection of texts. Implementation is achieved via a Python library back end and a web application front end built with React and D3.js for visualizations. TopEx has been successfully used to identify themes, topics and key words in a variety of corpora, including Coronavirus disease 2019 (COVID-19) discharge summaries and tweets. Feedback from the BioCreative VII Challenge Track 4 concludes that TopEx is a useful tool for text exploration for a variety of users and tasks.
Introduction
In this digital age, research in any field inevitably requires the analysis of large data sets, including textual data. It is important to become familiar with any new set of data prior to running an analysis (1), and this is especially true with textual data. Topic modeling can be used to explore a set of documents (a corpus) by identifying various topics contained in the corpus. Latent Dirichlet allocation (LDA) is a statistical topic modeling algorithm that assumes each document is composed of multiple topics and identifies the proportion of each topic in each document where a ‘topic’ is defined as a probability distribution over words (2). LDA is by far the most popular topic modeling approach, but there are many others including relational topic models, time-based topic models, topic models optimized for short texts and others (3). However, there are few applications that allow one to perform a topic analysis without requiring programming skills or knowledge of natural language processing (NLP) techniques. For example, Python packages, such as PyLDAvis (4) and LDAExplore (5), provide functions to perform a topic analysis on a set of texts as well as visualize the results; however, these tools require a knowledge of Python programming (1). There are applications that utilize graphical user interfaces, but many of these require a subscription (ATLAS.ti), are difficult to install/customize (6) (https://github.com/uwgraphics/SerendipSlim) or are no longer available (7–9). Thus, the analysis of unstructured text documents still poses challenges to many; hence, there is a need for an easy-to-use, programming-free topic exploration tool that can be utilized by researchers from any domain.
In this work, we present TopEx, an NLP tool that allows for easy exploration of topics in a set of text documents. TopEx is domain agnostic and is designed to allow processing and exploration of niche corpora, such as those associated with Coronavirus disease 2019 (COVID-19). With a user-friendly web interface and interactive graphical display of results, TopEx removes the barrier of having to learn a programming language or NLP techniques in order to explore topics present in a set of text documents, expanding the range of research nontechnical researchers can perform.
TopEx NLP pipeline
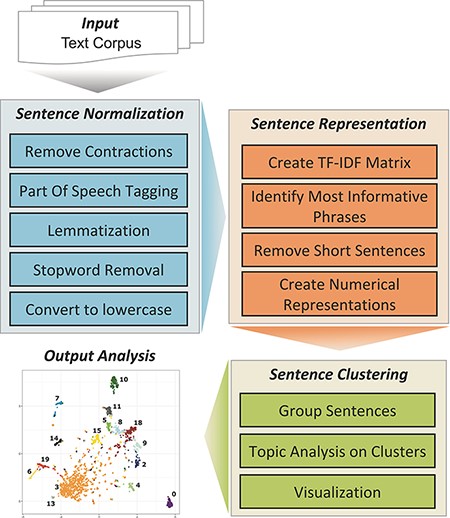
The TopEx algorithm is described in detail in the work by Olex et al. (10). The NLP pipeline implemented in TopEx is composed of three primary steps (Figure 1). At a high level, TopEx assumes that each sentence expresses one topic and aims to group sentences based on their similarity followed by identifying the main topic of each sentence group. This is done by first normalizing sentences (Sentence Normalization), which includes removing contractions and uninformative words (stopwords), identifying parts of speech, reducing words to their base form and converting all characters to lowercase. Stopwords are words that are not useful to the task at hand, such as ‘the’, ‘a’ or ‘by’ but can also include domain-specific words like ‘patient’. For example, the word ‘patient’ is generally not helpful when processing medical documents because all documents are about a patient, so it is not a word that is helpful when included in a topic. Second, sentences are converted into a numerical representation (Sentence Representation) by generating a term frequency-inverse document frequency (TF-IDF) matrix (11), which is created using the input set of texts (aka corpus). The TF-IDF is first used to identify each sentence’s ‘most informative phrase’, and then, a reduced form of the TF-IDF matrix is used to convert the phrase into a numerical vector that represents the whole sentence. TopEx defines the ‘most informative phrase’ as the phrase containing the six consecutive words (after Sentence Normalization) that achieve the highest part-of-speech-weighted sum of values in the TF-IDF plus a sentiment value [see the work by Olex et al. (10) for details]. Users can change the window for the number of consecutive tokens and can also choose to omit the sentiment score for creating the sentence representations. Third, the numerical sentence representations from all sentences in the corpus are pooled before clustering is performed on the numerical sentence vectors (Sentence Clustering) to group similar sentences together, and a topic analysis using LDA (2) is run on each cluster to identify main topics. LDA was chosen due to its popularity in the NLP field; however, other methods could be investigated for implementation in TopEx. By default, the TF-IDF matrix is created using the input corpus; however, users can optionally import custom background corpora to add additional domain-specific context to the analysis. Traditionally, LDA is run on an entire corpus and identifies multiple topics per document; however, it is not possible to identify which topics came from which sentences, or which topics are more cohesive than others. By first clustering the sentences on similarity, TopEx is able to provide the user a means to identify which sentences produce a topic and allows them to quickly verify by skimming clustered sentences manually instead of reviewing the entire corpus. This is advantageous as neither TopEx nor LDA identifies topic negations, which can be identified through manual review. In addition, the visualization of sentence similarity through scatter plots in TopEx allows users to identify which topics are more cohesive and distinct. Finally, results are visualized using word clouds and a scatter plot and can be saved in a delimited text file for easy browsing of sentence clusters. Running TopEx multiple times and viewing the word clouds to identify dominant topic words that are uninformative to the task at hand can aid in the identification of domain-specific stopwords.
Implementation and availability
The functionality of TopEx can be accessed through two modalities: (i) a Python 3.x library that can be downloaded from PyPI (https://pypi.org/project/topex) using the ‘pip install topex’ command and (ii) a web application with a graphical user interface (TopExApp). The TopEx Python library allows technical users the ability to integrate TopEx analyses into their NLP pipelines, as well as access the latest features and functionality. This modality is recommended for users with very large data sets. TopExApp is a web application with a graphical user interface that removes the high technical barrier traditionally associated with NLP and empowers researchers of any domain to directly utilize NLP tools without technical assistance. The TopExApp front end is built with the React JavaScript framework and the D3.js visualization library. The back end consists of a Python Flask Application Programming Interface (API). TopExApp is hosted on a web server at topex.cctr.vcu.edu for general public use on nonsensitive data. For users exploring documents containing protected health information, or otherwise sensitive data, a local Docker image can be built from the code on GitHub (https://github.com/VCUWrightCenter/TopExApp). Additionally, the source code for the TopEx Python library is also on GitHub (https://github.com/VCUWrightCenter/TopEx).
User interface and results exploration
The TopExApp web interface (Figure 2) allows users to load their input corpus in multiple formats from the ‘Load Data’ tab (Figure 2, first panel). These include (i) a set of text files, (ii) a pipe-delimited file containing the text of one document per line, (iii) an Excel file, (iv) a MEDLINE formatted file or (v) by entering key words to search PubMed for relevant abstracts directly from the application. Experienced users can customize the NLP pipeline by changing default settings in the ‘Advanced Parameters’ section of the ‘Parameters’ tab (Figure 2, second panel top and third panel); however, the default settings should produce good results for many analyses. The Parameters tab is organized into basic and advanced parameters, with the advanced parameters hidden in a dropdown box. Basic parameters are those frequently used to perform a TopEx analysis, which includes changing the number of clusters the sentences are grouped into and adding a stopwords file. Advanced parameters include additional sentence embedding, clustering and visualization parameters (default settings shown in Figure 2, third panel). It is recommended that users read the manual and have some basic understanding of NLP algorithms before changing the advanced parameters.

Screenshots of TopEx tabs and menu items. First panel shows the ‘Load Data’ tab with options for importing text data into TopEx. Second Panel shows the ‘Parameters’ (top), ‘Re-Cluster’ (middle) and ‘Import/Export’ (bottom) tabs. ‘Parameters’ tab allows customization of analysis and algorithm settings. ‘Re-Cluster’ enables quick adjustment of the cluster number without re-running the NLP pipeline from scratch. ‘Import/Export’ enables saving TopEx results or importing previous TopEx analyses. Third panel expands the ‘Advanced Parameters’ section of the ‘Parameters’ tab.
Results are visualized as a scatter plot and a set of word clouds. By default, the scatter plot is generated using the uniform manifold approximation and projection (UMAP) dimensionality reduction algorithm (12). However, users can change the algorithm used to create this plot in the Advanced Parameters drop-down, which includes options for t-distributed stochastic neighbor embedding (tSNE) (13), singular value decomposition and multidimensional scaling. All of these methods take the highly dimensional numerical matrix of sentence embeddings and reduce it to two dimensions so that it can be easily visualized as a scatter plot (Figure 3, tSNE layout; Figure 4, UMAP layout). Each point represents a sentence, and when using the UMAP or tSNE projections, points that are closer together indicate sentences that discuss similar topics. tSNE works well for corpora that have a lot of highly similar or identical sentences, such as documents that contain templated sentences, because it will not directly plot points on top of each other like UMAP does. The UMAP visualization works well for corpora with diverse sentences as it creates a more condensed visualization. While singular value decomposition and multidimensional scaling work well as vectorization methods, they tend to not produce useful visualizations; however, they are provided as they are classic algorithms utilized in the NLP field. Ultimately, the choice of visualization is up to the end user and generally depends on personal preferences and usefulness in exploration of a corpus; thus, the authors recommend users experiment with this option. Regardless of the visualization method chosen, the color of the dot refers to the cluster that each sentence was assigned. Because clustering is performed in the high-dimensional space of the embeddings, sentence clusters may not directly correlate with their position on the scatter plot. Users can explore the data by hovering over points to see (i) the full text of the sentence, (ii) the most informative phrase in that sentence and (iii) a list of terms characterizing themes present in that sentence’s cluster (Figure 3, right side). In addition, word clouds of each cluster visualize the frequency of each word in a cluster as the size of the bubble (Figure 4, insets). Word clouds give the user insight into which words may be dominating the topic of a particular cluster. The raw data behind the visualizations, along with the row-level results of the topic and clustering analysis, can be downloaded using the Import/Export tab for further analysis and can be used in external programs such as R to generate high-quality, customizable figures (Figure 4).
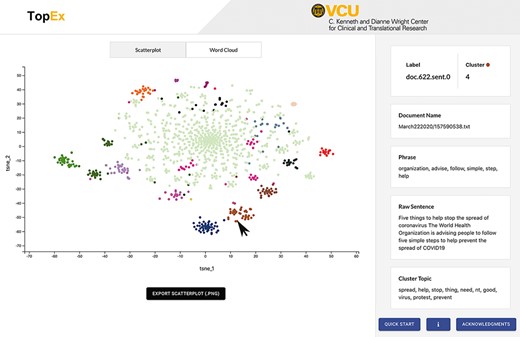
Screenshot of the TopEx interface showing results presented by a tSNE scatter plot and the sentence information displayed on the right when hovering over a point. Corpus used is a randomly sampled set of tweets from March 2020 in the COVID-19 Twitter Chatter data set discussed in the ‘Use Case’ section (same set of tweets that produced the UMAP visualization of Figure 4A).
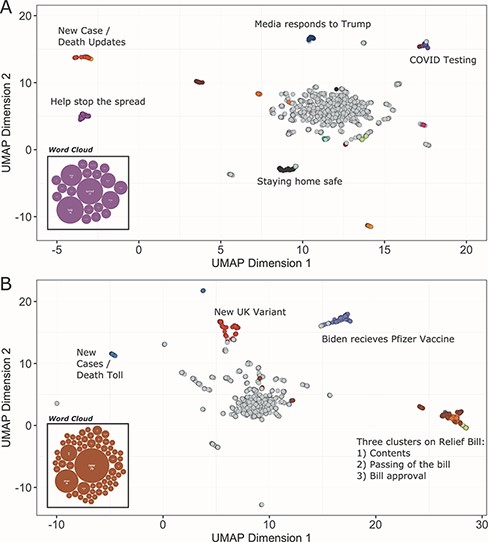
UMAP scatter plots and example word clouds from TopEx results for tweets from (A) March 2020 and (B) December 2020. Scatter plots were generated in R from the coordinate text file output by TopEx.
System performance
TopEx was built to be an exploratory tool, so there are no specific benchmarks that can give an estimate of its performance other than the interpretability and relevance of the results. However, for the initial use case, we did create a manually classified set of responses to assess whether or not TopEx could recreate those clusters (10). Briefly, the original TopEx use case was to identify common challenges experienced by fourth-year medical students participating in an acting internship through analyzing their written reflections in response to a guiding question and submitted via blog post. The development corpus used for algorithm evaluation contained 14 manually selected responses as they contained known challenges experienced by the students. A total of 172 sentences were annotated by two medical educators and one NLP expert as discussing one of the following topics: Confidence, Feeling Overwhelmed, Supportive Environment, System Issues or none. The test corpus was a randomly selected set of responses from a single month. Inter-annotator agreement (Cohen’s kappa statistic) averaged 0.56 for development and 0.51 for the testing corpus. These increased to 0.71 and 0.70, respectively, when only Supportive Environment and System Issues categories were considered. It was difficult to obtain good inter-annotator agreement while building the annotated reference data set, especially when annotating topics that were subjective, such as Feeling Overwhelmed and Confidence, which indicates that assigning sentences to predefined topics is even difficult for people. Qualitatively comparing the terms present in the four topics revealed that TopEx seemed to perform better on topics that had a distinct vocabulary, like the vocabulary around using a computer to place orders (10), and these were also the topics with a higher inter-annotator agreement (i.e. Supportive Environment and System Issues).
Example use case: evolution of COVID-19 pandemic tweets
TopEx is a versatile tool and can be used with any type of textual data. Table 1 provides a list of current use cases, both for internal operations (processing discharge summaries) and research. For this work, we provide another example use case that utilizes TopEx to explore the evolution of topics in a subset of COVID-related tweets during the year 2020. Briefly, we randomly sampled 2000 English language tweets from the 22nd day of each month starting on 22 March 2020 from the COVID-19 Twitter Chatter data set collected by Banda et al. (14). Figure 4 contains the results generated from clustering this subset of COVID tweets for March 2020 (Figure 4A) compared to December 2020 (Figure 4B) with topics of various clusters highlighted. Topics in March revolved around stopping the spread of COVID, staying home, COVID testing and the media’s reaction to then President Donald Trump. In December, the topics shifted to the new UK variant, vaccinations and the COVID relief bill. In both months, we see a cluster reporting new cases and the death toll. This cluster appeared in every month analyzed from March to December, which demonstrates that TopEx is able to identify consistent themes occurring throughout the year, as well as transient issues of interest to the community.
| Text type . | Use case . |
|---|---|
| Reflective medical writings | Identify common challenges experienced by medical students (Olex et al. (10) and second manuscript is under review). |
| COVID-19 discharge summaries | Identify key phrases and terms associated with COVID-19 patients to develop better rule-based queries using an in-house NLP system at VCU Massey Cancer Center. |
| Government COVID-19 communications | Identify how mitigation strategies implemented in South Korea changed over time during the first wave of the COVID-19 pandemic [poster presented at 43rd Annual Meeting and Scientific Sessions of the Society of Behavioral Medicine (15)]. |
| COVID-19 tweets | Assess changing topics of community interest during the pandemic (this manuscript). |
| Medical student narrative assessments | Identifying common themes in positive and negative feedback from formal assessments for medical students (two posters accepted to AAMC 2022 Annual Meeting and manuscript in preparation). |
| Text type . | Use case . |
|---|---|
| Reflective medical writings | Identify common challenges experienced by medical students (Olex et al. (10) and second manuscript is under review). |
| COVID-19 discharge summaries | Identify key phrases and terms associated with COVID-19 patients to develop better rule-based queries using an in-house NLP system at VCU Massey Cancer Center. |
| Government COVID-19 communications | Identify how mitigation strategies implemented in South Korea changed over time during the first wave of the COVID-19 pandemic [poster presented at 43rd Annual Meeting and Scientific Sessions of the Society of Behavioral Medicine (15)]. |
| COVID-19 tweets | Assess changing topics of community interest during the pandemic (this manuscript). |
| Medical student narrative assessments | Identifying common themes in positive and negative feedback from formal assessments for medical students (two posters accepted to AAMC 2022 Annual Meeting and manuscript in preparation). |
| Text type . | Use case . |
|---|---|
| Reflective medical writings | Identify common challenges experienced by medical students (Olex et al. (10) and second manuscript is under review). |
| COVID-19 discharge summaries | Identify key phrases and terms associated with COVID-19 patients to develop better rule-based queries using an in-house NLP system at VCU Massey Cancer Center. |
| Government COVID-19 communications | Identify how mitigation strategies implemented in South Korea changed over time during the first wave of the COVID-19 pandemic [poster presented at 43rd Annual Meeting and Scientific Sessions of the Society of Behavioral Medicine (15)]. |
| COVID-19 tweets | Assess changing topics of community interest during the pandemic (this manuscript). |
| Medical student narrative assessments | Identifying common themes in positive and negative feedback from formal assessments for medical students (two posters accepted to AAMC 2022 Annual Meeting and manuscript in preparation). |
| Text type . | Use case . |
|---|---|
| Reflective medical writings | Identify common challenges experienced by medical students (Olex et al. (10) and second manuscript is under review). |
| COVID-19 discharge summaries | Identify key phrases and terms associated with COVID-19 patients to develop better rule-based queries using an in-house NLP system at VCU Massey Cancer Center. |
| Government COVID-19 communications | Identify how mitigation strategies implemented in South Korea changed over time during the first wave of the COVID-19 pandemic [poster presented at 43rd Annual Meeting and Scientific Sessions of the Society of Behavioral Medicine (15)]. |
| COVID-19 tweets | Assess changing topics of community interest during the pandemic (this manuscript). |
| Medical student narrative assessments | Identifying common themes in positive and negative feedback from formal assessments for medical students (two posters accepted to AAMC 2022 Annual Meeting and manuscript in preparation). |
BioCreative user feedback
TopEx was submitted to the BioCreative VII Challenge Track 4: COVID-19 Text Mining Tool Interactive Demo (16) and was tested by a variety of users. As the system developers, we provided four target-audience users (three nontechnical clinical researchers and one grant funding organization representative), and the Track 4 organizers solicited the research community for additional participants. Users were sent a tutorial to complete and then asked to test out the system with some of their own data. Feedback was obtained from a survey of 30 questions developed by the BioCreative Track 4 organizers with 14 requiring a numerical rating and 16 asking for unstructured text feedback. Table 2 lists the questions that required a numerical rating. The BioCreative column provides the average score TopEx received from users as part of the BioCreative Challenge, both with and without a single outlier from someone who was looking for a gene disease association tool rather than topic trends. TopEx received a total of seven responses during the BioCreative Challenge evaluation with two who indicated they are directly involved in COVID-19 research. Two users were from government organizations, one from a patient organization that funds research grants and the remaining four from an academic setting. Users indicated that their field of work includes biocuration, informatics and grant funding. Prior to this challenge, only two users had used topic analysis systems in the past, which included topic modeling using LDA and Chalklabs services. The following sections include a summary of the unstructured response survey questions, which include tested functionalities, bottlenecks, usefulness of results and whether the evaluator would consider using the system in the future.
User survey questions with numerical ratings and the average score for TopEx during the first (BioCreative, n = 7) and second (post-BioCreative, n = 6) rounds of evaluation with and without one outlier each (n = 6 and n = 5, respectively)
| Question . | Rating rubric . | TopEx Score from BioCreative (with outlier) . | TopEx Score Post-BioCreative (with outlier) . |
|---|---|---|---|
| I think that I would like to use this system frequently. | 3.2 (2.9) | 3 (2.8) | |
| I found the system unnecessarily complex. | 2.3 (2.4) | 2 (2.3) | |
| I thought the system was easy to use. | 3 (3) | 4.2 (3.8) | |
| I think I would need support from the developer to be able to use this system. | 3.3 (3.1) | 2.8 (3.2) | |
| I found the various functions of the system well integrated. | 3.8 (3.7) | 3.8 (3.5) | |
| I thought there was too much inconsistency in this system. | 1 = strongly disagree | 2.3 (2.4) | 1.6 (2) |
| I would imagine that most people would learn to use this system very quickly. | 5 = strongly agree | 3.2 (3.1) | Not asked |
| I found the system very cumbersome to use. | 2.8 (2.9) | 1.8 (2.2) | |
| The system has met my expectations. | 3.2 (3.1) | 4 (3.5) | |
| I felt very confident using the system. | 3.5 (3.1) | 3.6 (3.2) | |
| I needed to learn a lot of things before I could get going with the system. | 3.3 (3.4) | 2.8 (3) | |
| How easy was it to format and input data into this tool? | 1 = not at all easy 5 = extremely easy | 3 (3) | 3.2 (3.2) |
| Please rate your overall impression with the system. | 1 = very negative 5 = very positive | 3.3 (3.3) | 3.6 (3.2) |
| How likely is it that you would recommend this system to a colleague performing COVID-19 related research? | 1 = not at all likely 10 = extremely likely | 6 (6) | 7.8 (6.8) |
| Question . | Rating rubric . | TopEx Score from BioCreative (with outlier) . | TopEx Score Post-BioCreative (with outlier) . |
|---|---|---|---|
| I think that I would like to use this system frequently. | 3.2 (2.9) | 3 (2.8) | |
| I found the system unnecessarily complex. | 2.3 (2.4) | 2 (2.3) | |
| I thought the system was easy to use. | 3 (3) | 4.2 (3.8) | |
| I think I would need support from the developer to be able to use this system. | 3.3 (3.1) | 2.8 (3.2) | |
| I found the various functions of the system well integrated. | 3.8 (3.7) | 3.8 (3.5) | |
| I thought there was too much inconsistency in this system. | 1 = strongly disagree | 2.3 (2.4) | 1.6 (2) |
| I would imagine that most people would learn to use this system very quickly. | 5 = strongly agree | 3.2 (3.1) | Not asked |
| I found the system very cumbersome to use. | 2.8 (2.9) | 1.8 (2.2) | |
| The system has met my expectations. | 3.2 (3.1) | 4 (3.5) | |
| I felt very confident using the system. | 3.5 (3.1) | 3.6 (3.2) | |
| I needed to learn a lot of things before I could get going with the system. | 3.3 (3.4) | 2.8 (3) | |
| How easy was it to format and input data into this tool? | 1 = not at all easy 5 = extremely easy | 3 (3) | 3.2 (3.2) |
| Please rate your overall impression with the system. | 1 = very negative 5 = very positive | 3.3 (3.3) | 3.6 (3.2) |
| How likely is it that you would recommend this system to a colleague performing COVID-19 related research? | 1 = not at all likely 10 = extremely likely | 6 (6) | 7.8 (6.8) |
User survey questions with numerical ratings and the average score for TopEx during the first (BioCreative, n = 7) and second (post-BioCreative, n = 6) rounds of evaluation with and without one outlier each (n = 6 and n = 5, respectively)
| Question . | Rating rubric . | TopEx Score from BioCreative (with outlier) . | TopEx Score Post-BioCreative (with outlier) . |
|---|---|---|---|
| I think that I would like to use this system frequently. | 3.2 (2.9) | 3 (2.8) | |
| I found the system unnecessarily complex. | 2.3 (2.4) | 2 (2.3) | |
| I thought the system was easy to use. | 3 (3) | 4.2 (3.8) | |
| I think I would need support from the developer to be able to use this system. | 3.3 (3.1) | 2.8 (3.2) | |
| I found the various functions of the system well integrated. | 3.8 (3.7) | 3.8 (3.5) | |
| I thought there was too much inconsistency in this system. | 1 = strongly disagree | 2.3 (2.4) | 1.6 (2) |
| I would imagine that most people would learn to use this system very quickly. | 5 = strongly agree | 3.2 (3.1) | Not asked |
| I found the system very cumbersome to use. | 2.8 (2.9) | 1.8 (2.2) | |
| The system has met my expectations. | 3.2 (3.1) | 4 (3.5) | |
| I felt very confident using the system. | 3.5 (3.1) | 3.6 (3.2) | |
| I needed to learn a lot of things before I could get going with the system. | 3.3 (3.4) | 2.8 (3) | |
| How easy was it to format and input data into this tool? | 1 = not at all easy 5 = extremely easy | 3 (3) | 3.2 (3.2) |
| Please rate your overall impression with the system. | 1 = very negative 5 = very positive | 3.3 (3.3) | 3.6 (3.2) |
| How likely is it that you would recommend this system to a colleague performing COVID-19 related research? | 1 = not at all likely 10 = extremely likely | 6 (6) | 7.8 (6.8) |
| Question . | Rating rubric . | TopEx Score from BioCreative (with outlier) . | TopEx Score Post-BioCreative (with outlier) . |
|---|---|---|---|
| I think that I would like to use this system frequently. | 3.2 (2.9) | 3 (2.8) | |
| I found the system unnecessarily complex. | 2.3 (2.4) | 2 (2.3) | |
| I thought the system was easy to use. | 3 (3) | 4.2 (3.8) | |
| I think I would need support from the developer to be able to use this system. | 3.3 (3.1) | 2.8 (3.2) | |
| I found the various functions of the system well integrated. | 3.8 (3.7) | 3.8 (3.5) | |
| I thought there was too much inconsistency in this system. | 1 = strongly disagree | 2.3 (2.4) | 1.6 (2) |
| I would imagine that most people would learn to use this system very quickly. | 5 = strongly agree | 3.2 (3.1) | Not asked |
| I found the system very cumbersome to use. | 2.8 (2.9) | 1.8 (2.2) | |
| The system has met my expectations. | 3.2 (3.1) | 4 (3.5) | |
| I felt very confident using the system. | 3.5 (3.1) | 3.6 (3.2) | |
| I needed to learn a lot of things before I could get going with the system. | 3.3 (3.4) | 2.8 (3) | |
| How easy was it to format and input data into this tool? | 1 = not at all easy 5 = extremely easy | 3 (3) | 3.2 (3.2) |
| Please rate your overall impression with the system. | 1 = very negative 5 = very positive | 3.3 (3.3) | 3.6 (3.2) |
| How likely is it that you would recommend this system to a colleague performing COVID-19 related research? | 1 = not at all likely 10 = extremely likely | 6 (6) | 7.8 (6.8) |
Tested functionalities
Functionalities tested by the users included uploading data in the different supported formats, modifying the parameters, clustering, visualizing and exporting results. The most common functionality tested was uploading data in a format other than the default of a set of text files; specifically, the Comma Separated Value (CSV) upload feature was mentioned by three users, and the PubMed search feature with queries like ‘Craniosynostosis AND gene AND variant’, ‘Short QT syndrome’, ‘COVID-19 AND gene AND variant’, ‘(ncRNA or miRNA) AND Alzheimer’, ‘long covid’ and ‘Glioblastoma’ was commented on by four users in the survey responses. In addition, users explored the visualizations by zooming in and hovering over the plots to see individual sentence information, and one user tested the re-clustering functionality.
Positive feedback and usefulness of results
The majority of users (five out of seven) specifically indicated that they liked the interface because it was ‘very clean’, ‘easy to use and navigate’, ‘intuitive and relatively easy to follow’ and ‘the look and feel are very appealing’. Additionally, users indicated ‘it is very quick in returning clustering’, useful for ‘identifying trends in the data’ and for obtaining a ‘quick grasp to understand topics’. One user noted that the hovering feature for viewing the details of a specific data point in the visualization was helpful, and another appreciated the multiple options for loading input data, particularly the PubMed search option. Overall, five out of seven users indicated that the returned results and program outputs were in a useful format.
Bottlenecks and suggested improvements
Four out of seven users identified bottlenecks when using the system. The most frequent was due to poor error handling when loading documents that were formatted improperly, followed by a lack of clarity on the meaning and purpose of the many customizable parameters provided by TopEx. Other users also reported long runtimes for large queries, specifically large numbers of manuscript or grant abstracts, which limited the utility of TopEx for use in their work.
The most frequently suggested improvement was to provide lay descriptions of the parameters for nonspecialist users. While it was noted by one user that the parameter explanation is extensive in the user documentation, it was suggested that some of this material be posted directly on the site to aid users in navigating the parameter refinement process. Additionally, clarifying the formatting requirements for CSV upload was suggested, as several users were unable to get this functionality to work. Other improvements include allowing users to enter stopwords into a text box without having to upload a separate file, including a pagination feature for the uploaded documents, so the scroll bar is not so long for large inputs, referencing the source document name (i.e. PubMed identifier (PMID) or file name) on the details drill down and in downloaded results, higher quality export of images, the ability to submit jobs and have the results emailed to you, incorporating named entity recognition (NER) and other user interface improvements.
Overall impression and future use
While a few users had difficulties, those that were able to use the system found it easy to navigate and intuitive with results being output in a helpful format for exploration. The majority of users indicated that they would either definitely or possibly consider using TopEx for their own research in the future. Those that would not use TopEx included one person who was looking for a different type of tool to do gene–disease associations and a second who experienced very long run times. TopEx scored an average of 3.3 for overall impression (Table 2). When asked if they would recommend TopEx to a colleague involved in COVID-19 research, all but two users rated TopEx with a score of 5 or greater (highest score of 8 and lowest score of 1). Thus, the overall impression was that they liked the clean interface, found the system simple to navigate and that the tool would be useful in future research, including COVID-19.
Future uses of the system included analyzing different types of textual data. The users described various domains in which the system could be useful for analyzing different types of data including thematic gene lists for curation, topic analysis of grant funders and interview transcripts. A summary of suggested use cases for TopEx is provided in Table 3.
| Text type . | Use case . |
|---|---|
| PubMed abstracts | Identify main themes in a set of queried abstracts from PubMed. |
| Grant summaries | Identify topics addressed in a set of grants that need to be assigned to reviewers. |
| Publications | Identify thematic gene lists for manual curation from a collection of publications. |
| Interview transcripts | Analysis of transcripts of interviews in social behavioral work for common themes. |
| Open-ended survey/blog responses | Assessing themes or topics addressed in open-ended survey responses or topic-focused blog posts. |
| Text type . | Use case . |
|---|---|
| PubMed abstracts | Identify main themes in a set of queried abstracts from PubMed. |
| Grant summaries | Identify topics addressed in a set of grants that need to be assigned to reviewers. |
| Publications | Identify thematic gene lists for manual curation from a collection of publications. |
| Interview transcripts | Analysis of transcripts of interviews in social behavioral work for common themes. |
| Open-ended survey/blog responses | Assessing themes or topics addressed in open-ended survey responses or topic-focused blog posts. |
| Text type . | Use case . |
|---|---|
| PubMed abstracts | Identify main themes in a set of queried abstracts from PubMed. |
| Grant summaries | Identify topics addressed in a set of grants that need to be assigned to reviewers. |
| Publications | Identify thematic gene lists for manual curation from a collection of publications. |
| Interview transcripts | Analysis of transcripts of interviews in social behavioral work for common themes. |
| Open-ended survey/blog responses | Assessing themes or topics addressed in open-ended survey responses or topic-focused blog posts. |
| Text type . | Use case . |
|---|---|
| PubMed abstracts | Identify main themes in a set of queried abstracts from PubMed. |
| Grant summaries | Identify topics addressed in a set of grants that need to be assigned to reviewers. |
| Publications | Identify thematic gene lists for manual curation from a collection of publications. |
| Interview transcripts | Analysis of transcripts of interviews in social behavioral work for common themes. |
| Open-ended survey/blog responses | Assessing themes or topics addressed in open-ended survey responses or topic-focused blog posts. |
Response to user feedback
After the BioCreative Challenge, the TopEx team worked to address some of the negative user feedback. The interface described in the previous sections, and shown in Figures 2 and 3, is the end result of these updates. Specifically, the interface and user’s manual were updated to improve clarity on import/export formats, and each parameter now has a tooltip that shows a brief description of the parameter when hovered over with the cursor. In addition, to help users get up and running quickly without having to read the extensive user’s manual, we have added a Quick Start Guide that can be accessed directly from the interface. Another critique was that there were a lot of parameters and it was difficult to identify which ones were the most important. To address this, we added an ‘Advanced Parameters’ drop-down that hides the majority of the parameter options, so the most important options are easy to find (Figure 2, third panel). Specifically, choosing the number of clusters and submitting a stopwords file are considered the primary parameters that most users should consider adjusting, so they were moved to the top and are always visible. When using the PubMed search option, users indicated it would be helpful to include the PMID in the output to identify the publications containing certain topics; thus, we added a column in the results output and a field on the interface that lists the PMID or original document name the sentence originated from. Finally, we added two new functionalities to TopEx: (i) the ability to import a native Microsoft Excel file and (ii) implementation of a biomedical NER toggle. The Excel file input option was added for user convenience as many data sets are in, or could be exported to, an Excel file format. Additionally, the NER option allows users to have their data set first run through the biomedical NER module, which merges multi-word biomedical terms into a single token that is not separated into individual words during the analysis. For example, the concept ‘breast cancer’ was originally broken into the words ‘breast’ and ‘cancer’; however, when the NER toggle has checked this, the term ‘breast cancer’ is kept together. The biomedical NER module is implemented using the ‘en_core_sci_sm’ model from SciSpacy (17).
Evaluation of improvements
After responding to user feedback, a second round of user evaluations was performed using a subset of questions from the original survey. The survey was sent out to both new and prior users. We received six responses where three had tested TopEx during the BioCreative Challenge and three were new users. The group of evaluators providing a second round of feedback contained a diverse set of users who work in biocuration, nursing, project management, advocacy and information technology. This round of feedback also contained an outlier who was a different user from the initial evaluation round. This user reviewed TopEx from an information technology point of view and scored very differently from the other evaluators who had backgrounds closer to the target user base. Table 2 shows scores with and without these outlier scores. Through the unstructured survey questions, users indicated that they tested various functionalities, including the PubMed search feature, Excel input format option, changing parameter settings, re-clustering, general navigation and ease of following the tutorial and user’s manual.
Positive feedback
Table 2 (post-BioCreative column) shows the average score TopEx received for each of the quantitative rating questions. As with the first round of evaluations, there was one user who provided divergent scores from the others for most questions; thus, average scores are shown with and without these outlier values. The majority of the users liked the interface and noted that it was ‘very visual’, ‘clean’ and ‘user friendly’, and several users commented that exporting the data was ‘very easy’, and it was ‘easy to get results quickly’. For the statement ‘I thought the system was easy to use’ (Table 2), TopEx received an increased score post-improvements to 4.2 (versus 3.0 pre-improvement), which was reduced to 3.8 with the outlier who stated that TopEx was ‘not user friendly’ for a ‘matured professor community’. In addition, most users disagreed with the statement ‘I found the system cumbersome to use’ with a reduction in the score post-improvement to 1.8 (2.2 with outlier) from a prior 2.8, indicating that the changes made to TopEx improved the usability of the system.
Users also liked the ability of TopEx to accept multiple input file formats, and when asked ‘How easy was it to format and input data into the tool?’ they rated TopEx at a 3.2, which is slightly increased from the previous rating of 3.0. Notably, getting data into the right format for any tool is a challenge, which can be exacerbated with NLP tools as text needs to be cleaned prior to analysis. TopEx is no exception and requires text data to be cleaned prior to analysis, which can be a challenge. Additionally, users commented that it was easy to change parameters and recluster and search using PubMed queries. Specifically, one user noted that they ‘love that you can search using PubMed queries and that the results from these include PMIDs as [they] use these to compare the results to manual abstract reviews [they have] done’. When asked ‘Were the data/results provided to you in an easy/useful format?’ two-thirds of users said yes, which is similar to the previous results, and one user specifically commented on the usefulness of the word cloud feature for exploring topics. Finally, when asked if they found any bottlenecks in the system, 87% of users said ‘no’ compared to 43% previously, and users gave TopEx a higher score when asked if it would be useful for COVID-19 research (7.8 versus a 6 previously).
Mixed and negative feedback
One of the issues from the BioCreative Challenge evaluation was the lack of documentation and integration of the documentation into the interface. When asked if users agreed or disagreed with the statement ‘I needed to learn a lot of things before I could get going with the system’, TopEx received lower score post-improvement of a 2.8 (3 with outlier) compared to a 3.3 indicating that this aspect was improved (i.e. users disagreed with the statement); however, feedback from comments was mixed. One user indicated ‘instructions were clear on the utilization’, while another commented that it was ‘unclear how to run [an] in-program PubMed query’. Additionally, a user commented that ‘It may help to provide a voice on the screen itself for the next step’, and another user stated they were ‘unclear on [the] interpretation of the results’. This mixed feedback indicates that some of the documentation issues were resolved; however, there is a need for more documentation on certain features, like the PubMed search options, and on how to interpret the results. In addition, adding more cues on the screen may help users navigate the analysis process better. Finally, one user suggested that providing ‘A synopsis of the intent of the TopEx tool and the benefits to the user’ on the home screen would be beneficial instead of having to find the user’s manual. Thus, while the documentation has improved, there is still room for further modifications that could improve the user’s experience.
Another major issue users identified was that the system would hang or crash when large data sets were uploaded, and there was a lack of useful error messages when the system did crash. Additionally, users noted that it would be helpful to have the ability to zoom in on the word clouds to see the smaller text, have the word clouds directly integrated with the scatter plot and that the colors chosen for some were hard to read (i.e. light green with white text). Finally, one user commented extensively on the interface functionality and highlighted certain areas that may be confusing to users, such as the ‘i’ button that contains the user’s manual, hamburger menu that hides the functional tabs and automatic enabling/disabling of the run button after performing certain operations.
Future work
Our goal for TopEx is to enable nontechnical researchers easy access to NLP analyses; thus, the user feedback both during and after the BioCreative Challenge has been very informative and helpful in identifying and prioritizing future improvements. Based on user feedback during the BioCreative Challenge, we added on-site documentation to aid users in navigating the various analysis parameters at their disposal, improved the functionality and documentation surrounding the input formats, added a new input format and added a biomedical NER toggle. After these changes, comments about the documentation shifted to the lack of documentation for certain features and the need for more documentation around the interpretation of results. Also, there was more focus on the program crashing with large inputs, as well as minor issues with the functionality for certain aspects of the user interface.
Our current top priority with TopEx is to improve run times on larger documents and to prevent the system from crashing with these inputs. At this time, it is recommended that users with large data sets utilize the TopEx Python library as the public server has limited space to build the required matrices needed for the analysis. An observed rule of thumb is to limit the analysis to less than 2000 documents with sizes averaging around a four-sentence paragraph each for the public version of TopEx. We have a few use cases that require a larger set of documents to be processed, but the current public implementation cannot handle this amount of data. At present, TopEx is best used on smaller documents such as tweets, feedback fields from surveys or other small text blobs. Additional future improvements include providing a text box so users may input stopwords within the application instead of uploading a text file, new functionality such as the implementation of different levels of clustering to include paragraph or document summarization along with sentence level clustering, implementation of other topic modeling approaches as well as improved visualizations and additional documentation, including short video tutorials.
Conclusions
TopEx is a novel, domain agnostic, NLP tool that provides a user-friendly interface for nontechnical users to explore topics present in a set of texts. It has already been shown to be useful in navigating reflective writings from medical students (10), COVID-19 discharge summaries and tweets. TopEx was submitted to the BioCreative VII Challenge Track 4 and was evaluated by a diverse group of users. Feedback was used to make improvements to TopEx before a second round of evaluations. The overall impression from users is that TopEx is easy and intuitive to navigate and provides useful output, and they would consider using TopEx in their future research. In conclusion, end users have indicated that TopEx is a user-friendly NLP tool that facilitates the exploration of topics in a set of texts and enhances the ability of nontechnical researchers to explore and analyze text data.
Acknowledgements
The authors would like to thank Sean Kortola, Aiden Meyers and Suzanne Prince for work in implementing the first prototype of TopExApp for their senior year capstone project as students of Virginia Commonwealth University (VCU’s) Computer Science Department. The authors would also like to thank the organizers of the BioCreative Challenge and all users who tested and provided feedback for TopEx, including Timothy Coetzee, William Cramer, Darryl Estrada-Zavala, Deborah DiazGranados and Padma Raju.
Funding
Clinical Translational Science Award (UL1TR002649) from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Conflict of interest
None declared.



