-
PDF
- Split View
-
Views
-
Cite
Cite
Pengyu Du, Yingli Chen, Qianzhong Li, Zhimin Gai, Hui Bai, Luqiang Zhang, Yuxian Liu, Yanni Cao, Yuanyuan Zhai, Wen Jin, CancerMHL: the database of integrating key DNA methylation, histone modifications and lncRNAs in cancer, Database, Volume 2024, 2024, baae029, https://doi.org/10.1093/database/baae029
Close - Share Icon Share
Abstract
The discovery of key epigenetic modifications in cancer is of great significance for the study of disease biomarkers. Through the mining of epigenetic modification data relevant to cancer, some researches on epigenetic modifications are accumulating. In order to make it easier to integrate the effects of key epigenetic modifications on the related cancers, we established CancerMHL (http://www.positionprediction.cn/), which provide key DNA methylation, histone modifications and lncRNAs as well as the effect of these key epigenetic modifications on gene expression in several cancers. To facilitate data retrieval, CancerMHL offers flexible query options and filters, allowing users to access specific key epigenetic modifications according to their own needs. In addition, based on the epigenetic modification data, three online prediction tools had been offered in CancerMHL for users. CancerMHL will be a useful resource platform for further exploring novel and potential biomarkers and therapeutic targets in cancer.
Database URL: http://www.positionprediction.cn/
Introduction
A large number of studies had found that epigenetic modifications play an important role in tumorigenesis (1–3). The abnormal changes of epigenetic modifications may lead to the alterations of genome structure and gene expression and tumorigenesis (4–6). Because epigenetic changes are reversible, the identification of key epigenetic modifications in cancer may be useful for the study of targeted epigenetic therapies (7).
In recent years, with the rapid increase of DNA methylation, histone modifications and lncRNA associated with cancer, some specific resources on DNA methylation, histone modifications, and lncRNAs in diseases have been made. For instance, MethHC 2.0 (8), DiseaseMeth 3.0 (9), MethyCancer (10), MethDB (11), Pancan-meQTL (12) and DDMGD (13) included DNA methylation data obtained from experiments. PubMeth (14) and MeInfoText (15) extracted information about cancer methylation by mining literature. MethMarkerDB focused on some especial genes and methylated regions in cancer (16). HHMD (17) and iHMS (18) were databases on histone modification data obtained from experiments. Lnc2Cancer 3.0 (19), LncRNADisease 3.0 (20), LncTarD 2.0 (21), Lnc2Meth (22), LnCeCell (23), LncSEA 2.0 (24) and lncRNASNP2 (25) integrated the disease-related lncRNA data supported by experiment and computation.
In our previous works, the changes of DNA methylation and histone modification patterns in functional regions were explored by analyzing the DNA methylation and histone modification data in liver cancer, breast cancer, colon cancer, lung cancer and chronic myelogenous leukemia. Based on these changes, the key methylation sites and histone modification regions were discovered in these cancers and the effect of the key epigenetic modification changes on gene expression were deeply discussed.
Based on these databases and our previous studies, it will be useful to construct the platform of the integrated epigenetic modification data. Therefore, we established CancerMHL in which the information related key DNA methylation, histone modifications and lncRNAs in several cancers can be easily obtained. It will provide a useful platform for exploring novel epi-biomarkers and therapeutic targets in the study of these cancers. The current version of CancerMHL collects key methylation sites, histone modifications and their regions as well as lncRNAs related to cancer. Each item includes the detailed annotation information of epigenetic modification, such as regions, sites and their effects on gene expression. Furthermore, based on our previous studies, CancerMHL collected the distribution patterns of differential DNA methylation and differential histone modifications in the various regions of genomes for different cancers. It also provides the several online tools of effective prediction algorithms and risk assessment models.
Data collection and database content
Data collection and processing
We extracted key DNA methylation and histone modification data in cancer based on the articles published by our research team (26–34). The main data include: (i) key differential histone modifications and their important regions in cancer, as well as their regulated target genes; (ii) key differential methylation sites in cancer and their regulated target genes; (iii) the effect of key epigenetic modifications on gene expression; (iv) the distribution changes of DNA methylation of up- and down-regulated genes in different functional regions; (v) the important differential expression genes; (vi) the important histone modification regions associated with the different types of gene expressions; (vii) the correlation of different histone modifications in up- and down-regulated genes. For their regulated target genes, we annotated gene types based on TSGene 2.0 (35), ONGene (36), COSMIC (37) and TAG (38) databases. Furthermore, we used the PubMed literature database to verify if these genes have been experimentally confirmed to influence cancer occurrence and development.
Many studies have found that DNA methylation, histone modifications and lncRNAs play a cooperative role in the regulation of gene expression (39–41). To facilitate such research, we searched the PubMed literature database using the keywords ‘cancer,’ ‘gene,’ and ‘lncRNA’. According to the following criteria: (i) lncRNAs related to human diseases and (ii) consistency in target genes regulated by lncRNAs with those regulated by methylation and histone modifications, we retrieved information about lncRNAs and their regulatory mechanisms on target genes. Based on the cooperative regulatory mechanisms of lncRNAs with other regulatory factors on target genes, we categorized lncRNA regulation into the following types: (i) co-regulation with DNA methylation; (ii) co-regulation with histone modifications; (iii) co-regulation with transcription factors; (iv) co-regulation with DNA methylation and histone modifications; and (v) lncRNAs independently regulate target genes.
Database statistics
The current version of CancerMHL contained key epigenetic modifications from 10 types of cancers, including key DNA methylation sites in different functional regions, the key regions of different histone modifications, and important lncRNAs participated in different co-regulations. The statistical results are shown in Tables 1, 2 and 3, respectively. Furthermore, CancerMHL provided the distribution patterns of DNA methylation and histone modification in different regions by mining epigenetic modification data related to cancer. It also provided two online risk assessment tools and an early diagnosis tool for the convenience of medical researchers.
| Disease . | Regions . | |||||||
|---|---|---|---|---|---|---|---|---|
| Breast cancer | Promoter | Enhancer | Exon | Intron | CGI | N_Shelf | N_Shore | S_Shore |
| 21 | 32 | 6 | 25 | 72 | 5 | 5 | 1 | |
| Hepatocellular carcinoma | Promoter | 5ʹUTR | exon | CGI | N_Shelf | N_Shore | S_Shore | |
| 12 | 2 | 7 | 2 | 1 | 8 | 2 | ||
| Colon adenocarcinoma | TSS200 | TSS1500 | body | CGI | N_Shore | S_Shore | 5ʹUTR | |
| 2 | 4 | 19 | 5 | 4 | 4 | 1 | ||
| Pan-cancer | TSS200 | TSS1500 | Body | CGI | ||||
| 1 | 2 | 2 | 1 | |||||
| Disease . | Regions . | |||||||
|---|---|---|---|---|---|---|---|---|
| Breast cancer | Promoter | Enhancer | Exon | Intron | CGI | N_Shelf | N_Shore | S_Shore |
| 21 | 32 | 6 | 25 | 72 | 5 | 5 | 1 | |
| Hepatocellular carcinoma | Promoter | 5ʹUTR | exon | CGI | N_Shelf | N_Shore | S_Shore | |
| 12 | 2 | 7 | 2 | 1 | 8 | 2 | ||
| Colon adenocarcinoma | TSS200 | TSS1500 | body | CGI | N_Shore | S_Shore | 5ʹUTR | |
| 2 | 4 | 19 | 5 | 4 | 4 | 1 | ||
| Pan-cancer | TSS200 | TSS1500 | Body | CGI | ||||
| 1 | 2 | 2 | 1 | |||||
Abbreviation: TSS200, 200 bp in the upstream of the transcription start site (TSS). TSS1500, 200–1500 bp in the upstream of the TSS.
| Disease . | Regions . | |||||||
|---|---|---|---|---|---|---|---|---|
| Breast cancer | Promoter | Enhancer | Exon | Intron | CGI | N_Shelf | N_Shore | S_Shore |
| 21 | 32 | 6 | 25 | 72 | 5 | 5 | 1 | |
| Hepatocellular carcinoma | Promoter | 5ʹUTR | exon | CGI | N_Shelf | N_Shore | S_Shore | |
| 12 | 2 | 7 | 2 | 1 | 8 | 2 | ||
| Colon adenocarcinoma | TSS200 | TSS1500 | body | CGI | N_Shore | S_Shore | 5ʹUTR | |
| 2 | 4 | 19 | 5 | 4 | 4 | 1 | ||
| Pan-cancer | TSS200 | TSS1500 | Body | CGI | ||||
| 1 | 2 | 2 | 1 | |||||
| Disease . | Regions . | |||||||
|---|---|---|---|---|---|---|---|---|
| Breast cancer | Promoter | Enhancer | Exon | Intron | CGI | N_Shelf | N_Shore | S_Shore |
| 21 | 32 | 6 | 25 | 72 | 5 | 5 | 1 | |
| Hepatocellular carcinoma | Promoter | 5ʹUTR | exon | CGI | N_Shelf | N_Shore | S_Shore | |
| 12 | 2 | 7 | 2 | 1 | 8 | 2 | ||
| Colon adenocarcinoma | TSS200 | TSS1500 | body | CGI | N_Shore | S_Shore | 5ʹUTR | |
| 2 | 4 | 19 | 5 | 4 | 4 | 1 | ||
| Pan-cancer | TSS200 | TSS1500 | Body | CGI | ||||
| 1 | 2 | 2 | 1 | |||||
Abbreviation: TSS200, 200 bp in the upstream of the transcription start site (TSS). TSS1500, 200–1500 bp in the upstream of the TSS.
| Disease . | Histone modifications . | ||||||
|---|---|---|---|---|---|---|---|
| Hepatocellular carcinoma | H3K4me1 | H3K4me2 | H3K4me3 | H3K27me3 | H3K79me2 | H3K9ac | H3K27ac |
| 2 | 8 | 22 | 5 | 5 | 18 | 28 | |
| Breast cancer | H3K36me3 | H3K79me2 | H3K9ac | H3K27ac | |||
| 3 | 3 | 6 | 6 | ||||
| Disease | Histone modifications | Disease | Histone modifications | Disease | Histone modifications | ||
| Colorectal cancer | H3K79me3 | Lung adenocarcinoma | H3K79me2 | Chronic myelogenous Leukemia | H3K36me3 | ||
| 20 | 12 | 6 | |||||
| Disease . | Histone modifications . | ||||||
|---|---|---|---|---|---|---|---|
| Hepatocellular carcinoma | H3K4me1 | H3K4me2 | H3K4me3 | H3K27me3 | H3K79me2 | H3K9ac | H3K27ac |
| 2 | 8 | 22 | 5 | 5 | 18 | 28 | |
| Breast cancer | H3K36me3 | H3K79me2 | H3K9ac | H3K27ac | |||
| 3 | 3 | 6 | 6 | ||||
| Disease | Histone modifications | Disease | Histone modifications | Disease | Histone modifications | ||
| Colorectal cancer | H3K79me3 | Lung adenocarcinoma | H3K79me2 | Chronic myelogenous Leukemia | H3K36me3 | ||
| 20 | 12 | 6 | |||||
| Disease . | Histone modifications . | ||||||
|---|---|---|---|---|---|---|---|
| Hepatocellular carcinoma | H3K4me1 | H3K4me2 | H3K4me3 | H3K27me3 | H3K79me2 | H3K9ac | H3K27ac |
| 2 | 8 | 22 | 5 | 5 | 18 | 28 | |
| Breast cancer | H3K36me3 | H3K79me2 | H3K9ac | H3K27ac | |||
| 3 | 3 | 6 | 6 | ||||
| Disease | Histone modifications | Disease | Histone modifications | Disease | Histone modifications | ||
| Colorectal cancer | H3K79me3 | Lung adenocarcinoma | H3K79me2 | Chronic myelogenous Leukemia | H3K36me3 | ||
| 20 | 12 | 6 | |||||
| Disease . | Histone modifications . | ||||||
|---|---|---|---|---|---|---|---|
| Hepatocellular carcinoma | H3K4me1 | H3K4me2 | H3K4me3 | H3K27me3 | H3K79me2 | H3K9ac | H3K27ac |
| 2 | 8 | 22 | 5 | 5 | 18 | 28 | |
| Breast cancer | H3K36me3 | H3K79me2 | H3K9ac | H3K27ac | |||
| 3 | 3 | 6 | 6 | ||||
| Disease | Histone modifications | Disease | Histone modifications | Disease | Histone modifications | ||
| Colorectal cancer | H3K79me3 | Lung adenocarcinoma | H3K79me2 | Chronic myelogenous Leukemia | H3K36me3 | ||
| 20 | 12 | 6 | |||||
| Disease . | Regulation types (numbers) . | ||||
|---|---|---|---|---|---|
| Hepatocellular carcinoma | lncRNA (28) | lncRNA-Meth (9) | lncRNA-HM (7) | lncRNA-TF (2) | lncRNA-Meth-HM (1) |
| Colorectal cancer | lncRNA (17) | lncRNA-Meth (4) | lncRNA-HM (2) | lncRNA-Meth-HM (2) | |
| Non-small cell lung carcinoma | lncRNA (20) | lncRNA-Meth (1) | lncRNA-HM (3) | lncRNA-TF (5) | |
| Disease | Regulation types (numbers) | Disease | Regulation types (numbers) | ||
| Breast cancer | lncRNA (9) | lncRNA-Meth (1) | Colon adenocarcinoma | lncRNA (2) | |
| Lung adenocarcinoma | lncRNA (7) | lncRNA-TF (1) | Lung cancer | lncRNA (2) | |
| Chronic myeloid leukemia | lncRNA (6) | lncRNA-Meth (2) | Lung squamous cell carcinoma | lncRNA (1) | |
| Disease . | Regulation types (numbers) . | ||||
|---|---|---|---|---|---|
| Hepatocellular carcinoma | lncRNA (28) | lncRNA-Meth (9) | lncRNA-HM (7) | lncRNA-TF (2) | lncRNA-Meth-HM (1) |
| Colorectal cancer | lncRNA (17) | lncRNA-Meth (4) | lncRNA-HM (2) | lncRNA-Meth-HM (2) | |
| Non-small cell lung carcinoma | lncRNA (20) | lncRNA-Meth (1) | lncRNA-HM (3) | lncRNA-TF (5) | |
| Disease | Regulation types (numbers) | Disease | Regulation types (numbers) | ||
| Breast cancer | lncRNA (9) | lncRNA-Meth (1) | Colon adenocarcinoma | lncRNA (2) | |
| Lung adenocarcinoma | lncRNA (7) | lncRNA-TF (1) | Lung cancer | lncRNA (2) | |
| Chronic myeloid leukemia | lncRNA (6) | lncRNA-Meth (2) | Lung squamous cell carcinoma | lncRNA (1) | |
Abbreviation: Meth, DNA methylation. HM, histone modification.
| Disease . | Regulation types (numbers) . | ||||
|---|---|---|---|---|---|
| Hepatocellular carcinoma | lncRNA (28) | lncRNA-Meth (9) | lncRNA-HM (7) | lncRNA-TF (2) | lncRNA-Meth-HM (1) |
| Colorectal cancer | lncRNA (17) | lncRNA-Meth (4) | lncRNA-HM (2) | lncRNA-Meth-HM (2) | |
| Non-small cell lung carcinoma | lncRNA (20) | lncRNA-Meth (1) | lncRNA-HM (3) | lncRNA-TF (5) | |
| Disease | Regulation types (numbers) | Disease | Regulation types (numbers) | ||
| Breast cancer | lncRNA (9) | lncRNA-Meth (1) | Colon adenocarcinoma | lncRNA (2) | |
| Lung adenocarcinoma | lncRNA (7) | lncRNA-TF (1) | Lung cancer | lncRNA (2) | |
| Chronic myeloid leukemia | lncRNA (6) | lncRNA-Meth (2) | Lung squamous cell carcinoma | lncRNA (1) | |
| Disease . | Regulation types (numbers) . | ||||
|---|---|---|---|---|---|
| Hepatocellular carcinoma | lncRNA (28) | lncRNA-Meth (9) | lncRNA-HM (7) | lncRNA-TF (2) | lncRNA-Meth-HM (1) |
| Colorectal cancer | lncRNA (17) | lncRNA-Meth (4) | lncRNA-HM (2) | lncRNA-Meth-HM (2) | |
| Non-small cell lung carcinoma | lncRNA (20) | lncRNA-Meth (1) | lncRNA-HM (3) | lncRNA-TF (5) | |
| Disease | Regulation types (numbers) | Disease | Regulation types (numbers) | ||
| Breast cancer | lncRNA (9) | lncRNA-Meth (1) | Colon adenocarcinoma | lncRNA (2) | |
| Lung adenocarcinoma | lncRNA (7) | lncRNA-TF (1) | Lung cancer | lncRNA (2) | |
| Chronic myeloid leukemia | lncRNA (6) | lncRNA-Meth (2) | Lung squamous cell carcinoma | lncRNA (1) | |
Abbreviation: Meth, DNA methylation. HM, histone modification.
Database usage and access
Web interface
CancerMHL offers a user-friendly web interface that enables user to browse and retrieve the information of epigenetic modifications in cancer. The CancerMHL website comprises three main modules, including (i) ‘Search’ for accessing the information about key epigenetic modifications in cancer, (ii) ‘Conclusion’ for obtaining the distribution patterns of DNA methylation and histone modifications in cancer and (iii) ‘Tools’ for using online tools for risk assessment and the early diagnosis of cancer patients. In addition, on the ‘Help’ page, CancerMHL provides a detailed tutorial for the usage of the database.
Using the search tool to retrieve the key epigenetic modifications in cancer
To access interesting key epigenetic modifications, users can query the database as follows. First, on the ‘Search’ page, users can filter data by using the type, disease and the location of the desired key epigenetic modifications (Figure 1A). Once selections are made, click the ‘Search’ button and the query results will be shown as a responsive table, with each item representing information about a key epigenetic modification. There is a ‘Details’ button at the end of each item, which will provide more comprehensive annotation information, including the visual representations of the effects of epigenetic modifications on gene expression, the location details of epigenetic modifications, information about genes and the distribution of signals in important histone modification regions (Figure 1B).
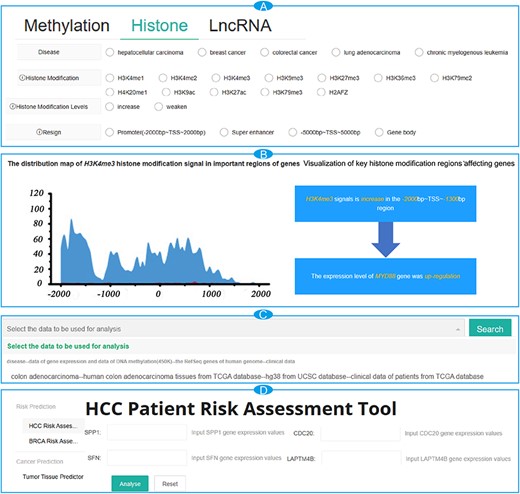
The interface of the browse module for CancerMHL. (A) The interface of the search modules; (B) search result page for MYD88 with detailed information; (C) module interface for analyzing results based on DNA methylation and histone modification data; (D) the interface of the tool modules.
Retrieving the distribution patterns of DNA methylation and histone modifications in cancer
On the ‘Conclusion’ page, users can explore the distribution patterns of DNA methylation and histone modifications, as well as some relevant important results, by selecting the data used for computational analysis from the dropdown list (Figure 1C).
Using of risk assessment tools and tumor tissue prediction tools
CancerMHL provides three online tools: a risk assessment tool for liver cancer patients, a risk assessment tool for breast cancer patients and a tumor tissue prediction tool. On the ‘Tools’ page, users select their interested tool from the options on the left side of the page. Following the instructions provided on the page, the users enter patient data into the input boxes (Figure 1D). Subsequently, by clicking the ‘Analyze’ button, users can obtain the patient’s risk grouping or predicted results.
Conclusions and future extensions
In this study, we established the integrated database of key epigenetic modifications in the several kinds of cancers. This platform provides a reference for further studying the effects of co-regulation with epigenetic modifications such as DNA methylation, histone modifications and lncRNAs on gene expression levels, including the effects of co-regulation with the different epigenetic modifications in same regions or same epigenetic modifications in different regions on gene expression, etc. Moreover, some of the important genes and epigenetic modification regions and sites can be used as potential epigenetic biomarkers. The establishment of this database will have significant guiding value for experimental researchers in exploring novel epigenetic biomarkers and therapeutic targets.
To make CancerMHL more comprehensive and useful, we will further improve and perfect the platform and database, and increase more annotation information and practical analysis tools. We plan to supplement relationship between chromatin accessibility annotation and key epigenetic modifications and the effects of epigenetic modifications on 3D genome structure in future versions.
Data availability
All the data can be downloaded from http://www.positionprediction.cn/.
Contribution statement
Y.C. and Q.L. provided comprehensive guidance; P.D. designed the database; H.B., L.Z., Y.L., Y.C., Y.Z. and W.J. provided the data; Z.G. collected and analyzed the data; P.D., Y.C. and Q.L. wrote the paper; all authors read and agreed the final manuscript.
Funding
National Natural Science Foundation of China [Nos 32160216, 62361047, 62161033].
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Zhiying Li and Yuanyuan Zhao for their contribution to some data collection.



