-
PDF
- Split View
-
Views
-
Cite
Cite
Susan M. Bello, Joel E. Richardson, Allan P. Davis, Thomas C. Wiegers, Carolyn J. Mattingly, Mary E. Dolan, Cynthia L. Smith, Judith A. Blake, Janan T. Eppig, Disease model curation improvements at Mouse Genome Informatics, Database, Volume 2012, 2012, bar063, https://doi.org/10.1093/database/bar063
Close - Share Icon Share
Abstract
Optimal curation of human diseases requires an ontology or structured vocabulary that contains terms familiar to end users, is robust enough to support multiple levels of annotation granularity, is limited to disease terms and is stable enough to avoid extensive reannotation following updates. At Mouse Genome Informatics (MGI), we currently use disease terms from Online Mendelian Inheritance in Man (OMIM) to curate mouse models of human disease. While OMIM provides highly detailed disease records that are familiar to many in the medical community, it lacks structure to support multilevel annotation. To improve disease annotation at MGI, we evaluated the merged Medical Subject Headings (MeSH) and OMIM disease vocabulary created by the Comparative Toxicogenomics Database (CTD) project. Overlaying MeSH onto OMIM provides hierarchical access to broad disease terms, a feature missing from the OMIM. We created an extended version of the vocabulary to meet the genetic disease-specific curation needs at MGI. Here we describe our evaluation of the CTD application, the extensions made by MGI and discuss the strengths and weaknesses of this approach.
Database URL:http://www.informatics.jax.org/
Introduction
The ability to curate disease-related data is imperative for many databases. There is a growing pool of available disease-related data, increasing interest from end users and pressure from funding agencies to make clear connections between the data from model organisms and the human diseases they reference. There are a number of disease vocabularies and ontologies that may be used for the purpose of annotating disease models each with its own advantages and disadvantages (1).
Background
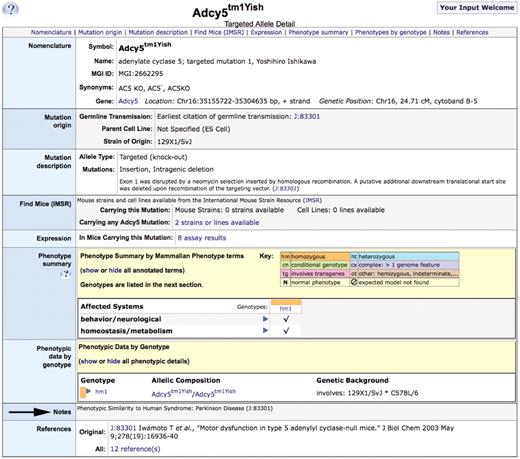
At Mouse Genome Informatics (MGI) (2), disease model annotations currently are made by associating specific mouse genotypes to Online Mendelian Inheritance in Man (OMIM) disease terms (OMIM.org). MGI filters the OMIM phenotype terms to exclude those that are not describing human diseases (e.g. HAIR MORPHOLOGY 2, 139 450) and loads only the OMIM disease terms. Annotations are made from published experimental-based assertions of the relationship between a mouse model and a human disease. These assertions often relate a mouse model to a general disease class. For example, Iwamoto et al. (3) stated ‘We have demonstrated that the disruption of the AC5 [official symbol ADCY5] gene led to a major deficit in AC activity in a striatal specific manner and an abnormal coordination […that] mimicked Parkinson's disease.’ While OMIM has over 20 different Parkinson disease sub-type records, there is no record for the general term Parkinson disease and none of the OMIM Parkinson disease records are associated with the human gene ADCY5. If OMIM had an ADCY5-specific Parkinson disease record, MGI could annotate this model to that record. However, we do not annotate etiologically distinct mouse models to an OMIM record unless the publication specifically refers to the exact disease. Thus, the model described by Iwamoto et al. cannot be annotated directly to any OMIM term. Such models are instead annotated in structured text fields (Figure 1). While text annotations allow users to view the model statements, the ability to search for and compute over these annotations is extremely limited. In addition, descriptions of complicated models involving multiple mutations on complex genetic backgrounds can be difficult to describe clearly in such structured text form. Further, because these text annotations are visually separated from the phenotypic annotations for the same model in the web display, connections between phenotype and disease may be difficult for users to identify.
Allele detail page for Adcy5tm1Yish, arrow indicates the structured text disease annotation in the ‘Notes’ section of the page.
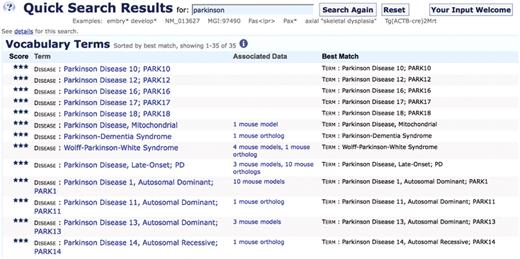
OMIM has been used by MGI for disease associations because of the presence of detailed disease descriptions, links between disease records and human genes and familiarity to biomedical researchers. However, the absence of hierarchical structure in OMIM means that there is no grouping mechanism beyond text searching to allow users to view all models of a disease such as Parkinson disease. Instead, users must collate models from each of the specific OMIM Parkinson disease records and models annotated in structured text in order to create a complete list (Figure 2). This situation is exacerbated as OMIM adds more records for specific types of a disease and the numbers of mouse models of human disease increase.
Partial search results from MGI for the keyword ‘Parkinson’. Users currently have no simple way to create a unified set of all mouse models of Parkinson disease.
Strategy
MGI sought to identify a disease ontology or vocabulary to improve curation of mouse models of human disease. General criteria for selecting a disease ontology have been defined previously (1, 4). The criteria considered most essential for annotation mouse models of human disease include several of those described by Bodenreider and Burgun (1) i.e. coverage of diseases, regular maintenance, support for reasoning and open availability. Additional criteria include stability of the vocabulary, percentage of terms with definitions, inclusion of synonyms and familiarity of the vocabulary to the user community. A final and necessary consideration for MGI curatorial representations is the incorporation of OMIM as part of the terminology.
These additional criteria are generally applicable to the use of any ontology. A stable ontology avoids the need for extensive and repeated recuration of data. Deep synonym coverage allows for easier identification of diseases from the literature and for more effective searching of the data by users. Definitions provide a description of the disease to aid in understanding of the disease term and provide a basis for comparison to the model. Familiarity of the user community improves the likelihood that users will readily find the disease representation they are seeking. In addition, for MGI, OMIM links were considered essential for the migration of existing annotations to the new vocabulary, to meet end user needs and to maintain access to the human disease to human gene annotations provided by OMIM.
Of the existing disease ontologies and vocabularies, two were identified as containing at least some links to OMIM; the Disease Ontology (DO) (5) and the MErged DIsease voCabulary (MEDIC) developed at the Comparative Toxicogenomics Database (CTD) (6). While the DO may grow into a better long-term solution, it was, at the time we undertook this evaluation, not nearly mature or robust enough to be useful for curating disease data. The DO was being extensively revised (which negatively impacts its stability), only 11% of the terms had definitions (as of 21 June 2010), and while OMIM IDs were being added, many were still missing and there was uneven mapping of OMIM diseases within the DO (Drs L. Schriml and W. Kibbe, personal communication). Therefore, we undertook an extensive evaluation of MEDIC.
CTD MEDIC
CTD created, implemented and maintained MEDIC, a disease vocabulary created by merging disease terms from OMIM with the disease subsections in Medical Subject Headings (MeSH) (Davis, AP et al., submitted for publication). Briefly, MeSH is a structured, hierachical thesaurus created and maintained by the National Library of Medicine to index journal articles (http://www.nlm.nih.gov/mesh/). Two subsections of MeSH were used to create the vocabulary: Diseases [C] and Mental Disorders [F03]. OMIM terms were limited to those with an associated National Center for Biotechnology Information accession ID (gene or locus). The merged vocabulary effectively is the MeSH hierarchy onto which all selected OMIM terms have been mapped based on lexical similarity or symptom matching for OMIM terms lacking a lexical match (i.e. OMIM 101 000, NEUROFIBROMATOSIS, TYPE II merged with MeSH D016518, Neurofibromatosis 2). Mapped OMIM terms were either merged with a MeSH term(s) or made a subterm (child) of a MeSH term(s). The use of symptom-based mappings allows for rapid and consistent mapping, but curation issues can result, and this is discussed more fully below.
CTD loads the vocabularies from MeSH and OMIM on a monthly basis and quality control processes identify changes to these vocabularies, which require curation of the merged vocabulary. This vocabulary is freely available from CTD and can be viewed on the web at http://ctd.mdibl.org/voc.go?type=disease.
Results and discussion
MEDIC was evaluated to determine its suitability for use in curation of mouse models of human disease by MGI. We considered the breadth and depth of disease terms in the vocabulary in relation to disease models in MGI. In addition, we considered the quality and consistency of the OMIM to MeSH mappings, the ability of the vocabulary to be modified to meet needs other than those for which it was originally created and ongoing maintenance requirements.
Breadth of coverage
Breadth of coverage refers to the extent to which an ontology covers a particular set of concepts. To determine the breadth of coverage, the full set of OMIM diseases in use at MGI was compared with the 4049 OMIM terms included in MEDIC at the time of analysis. This analysis was conducted twice. The first analysis defined OMIM terms used by MGI as any OMIM term loaded into MGI, regardless of whether or not the term had any associated mouse model or mouse gene. The first analysis, conducted in June 2010, identified 347 OMIM terms in MGI, which were absent from MEDIC. Two hundred and fifty-nine of these were in CTD's set of OMIM terms, which had been reviewed but not mapped to any MeSH term. Of these 259, 214 were determined to represent phenotypes or unmapped genes and not diseases. As a result, these OMIM terms were excluded from the set of OMIM terms displayed in MGI. About 30 of the 259 were either chromosome aberration syndromes (29) or diseases (1) with only very general symptom descriptions. These were determined to be of low priority based on the presumed low probability of the development of mouse models for these diseases and therefore left unmapped. An advantage of this vocabulary is the ease with which these OMIM terms could be added if a mouse model was ever identified. The final 15 OMIM terms in the unmapped set of 259 were mapped to MeSH terms. The remaining 88 terms from the original 347 were either new OMIM terms or OMIM terms without an associated gene (which were not part of the initial objectives of the vocabulary). All of these were individually mapped to at least one MeSH term, in an updated version of the vocabulary. From this first analysis then only 103 OMIM terms (15 plus 88) necessary for MGI curation were missing from the 4049 OMIM terms in MEDIC at the time of analysis, representing a deficiency in breadth of coverage of only 2.5% (103/4049).
A second analysis, conducted in August 2010, defined OMIM terms used in MGI as terms with either an associated mouse model or mouse gene. This analysis identified an additional 212 terms in MGI but absent from MEDIC. Of the 212, 37 were repeats from the first analysis. These 37 were all terms that had been rejected in the first analysis either as low priority unmapped terms or terms that should be excluded. Of the remaining 175, 90 were new OMIM terms that had not yet been mapped and the remaining 85 were existing OMIM terms without an associated gene (which were not part of the initial objectives of the vocabulary). All 175 unmapped OMIM terms were then examined and either mapped to appropriate MeSH terms or added to the unmapped term set. Of the 175, 12 were identified as not being disease terms and placed in the unmapped term set. The remaining 163 were individually mapped to at least one MeSH term. All additional mappings were added to an updated version of the vocabulary. In this second analysis then only 85 OMIM terms necessary for MGI curation were found missing, again representing a small deficiency in breadth of coverage.
Both analyses determined that CTD's scope for MEDIC, OMIM disease terms with an associated human gene, was not sufficient to meet all of MGI's disease curation needs. However, the additional terms needed could be readily identified and the creation of the additional MeSH mappings will require minimal periodic MGI curator time (around one curator day per quarterly update).
Depth of coverage
Depth of coverage refers to the precision of the vocabulary terms, or the level of detail (specificity) within an ontology. As MEDIC was originally created, OMIM terms that were of the type ‘Disease Name #’ (e.g. AGAMMAGLOBULINEMIA 1, 601 495; AGAMMAGLOBULINEMIA 6, 612 692) were merged into the generic MeSH term for that disease (e.g. Agammaglobulinemia, D000361). This compression of the more specific disease terms is undesirable at MGI where a distinction is defined between mouse models with similar or differing etiology compared to the human disease.
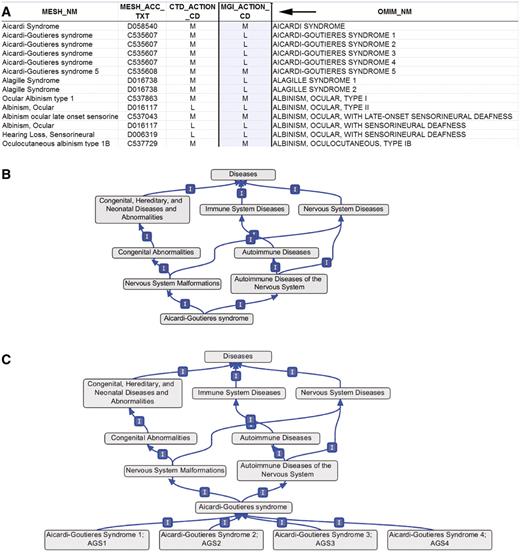
Again the vocabulary proved to be easily modified to meet MGI's needs. The mappings of OMIM to MeSH are maintained with a field indicating whether an OMIM term should be merged with (M) or made a child of (L) a MeSH term. An MGI-specific field (MGI_Action_CD) was added to allow for differing levels of term specificity. For example, in Figure 3A the MGI field specifies that the OMIM terms Alagille syndrome 1 and Alagille syndrome 2 should be made children of the MeSH term Alagille syndrome. The CTD field specifies that the same terms should be merged with Alagille syndrome. Similarly in CTD, the OMIM terms Aicardi–Goutieres syndromes 1–4 are merged with the MeSH term Aicardi–Goutieres syndrome (Figure 3B), while in MGI the OMIM terms are made children of the MeSH term (Figure 3C). However, in both the MGI and CTD versions the OMIM term Aicardi–Goutieres syndrome 5 is merged with the lexically identical MeSH term Aicardi–Goutieres syndrome 5. In all, MGI required approximately 740 terms to be added as children of MeSH terms where CTD had merged the OMIM term into the MeSH term. This difference resulted in creation of an MGI-specific variant of MEDIC. Both versions contain the same terms and differ only in the merge/child organizational structure described above. The extended version of the vocabulary is available in Open Biomedical Ontology (OBO) format at ftp://ftp.informatics.jax.org/pub/mosh. As MeSH does not use defined relationships between terms, the OBO-formatted file was created assuming all relationships are ‘is_a’ relationships.
(A) Section of the OMIM to MeSH mapping spreadsheet. Arrow indicates the MGI-specific field (MGI_Action_CD) used to generate the extended version of MEDIC. M, merge; L, leaf. (B) Graphical display of the OMIM terms Aicardi–Goutieres syndromes 1–4 within MEDIC, all four OMIM terms are merged with the MeSH term Aicardi–Goutieres syndrome. (C) Graphical display of the OMIM terms Aicardi–Goutieres syndromes 1–4 within MEDIC as used at MGI, all four OMIM terms are child terms to the MeSH term Aicardi–Goutieres syndrome.
Mapping consistency and quality
Most OMIM terms are readily mapped to MeSH terms based on lexical similarity. For example AGAMMAGLOBULINEMIA 1 (601 495) maps to the MeSH term Agammaglobulinemia (D000361) and PARKINSON DISEASE, LATE-ONSET (168 600) maps to the MeSH term Parkinson Disease (D010300). These mappings are all of high quality and highly consistent. These lexical mappings constitute the majority of the OMIM to MeSH mappings. Many of the OMIM terms that do not have a good lexical match in MeSH are for complex syndromes. Disease symptoms are identified from all available information in OMIM, e.g. clinical synopses, disease descriptions. By adopting a straightforward mapping of symptom to disease class, a high level of consistency can be maintained for these mappings. For example, the clinical synopsis for the disease OCULOAURICULAR SYNDROME (OMIM 612109) lists symptoms involving the subcategories ears and eyes. Therefore, this disease is mapped to the MeSH terms Ear Diseases (D004427) and Eye Abnormalities (D005124). In addition, for syndromes with less informative names symptom-based mapping may be informative for users. For example, mapping the OMIM term RIDDLE SYNDROME (611 943) to the MeSH terms for its symptoms (immune deficiency syndromes, learning disorders and facies) provides insights into the disease.
Not all symptom-based mappings are as straight forward as that of OCULOAURICULAR SYNDROME. There are two main pitfalls of symptom-based mappings. First, because a disease may produce a symptom in an organ or tissue it does not necessarily mean that all types of that disease are a disease of that organ or tissue. For example, in MeSH, albinism is a child of eye diseases and pigmentation diseases, while experts would agree that albinism is a pigmentation disease, not all forms of albinism are eye diseases. For example, piebaldism is a child of albinism and therefore a child of eye diseases but does not have an eye phenotype. Second, some symptom descriptions may lead to erroneous mappings if the mapping is not constructed or reviewed by an expert clinician. Symptoms described as being ‘like’ some other disease or syndrome, may be lexically, yet erroneously, mapped to that disease. For example, patients with Lujan–Fryns syndrome are described as having ‘Marfanoid habitus’, a term lexically related to the term ‘Marfan’ but whose definition is not related to Marfan syndrome. The symptom-based association assertion results in a mapping of Lujan–Fyrns syndrome to Marfan syndrome, which is incorrect. These kinds of situations require experts in disease phenotypes to identify, review and curate. Such clinical experts must be an integral part of any disease ontology development effort.
Despite these potential pitfalls, the vast majority of the OMIM to MeSH mappings in MEDIC were found to be highly consistent and of very good quality. In addition, as MeSH adds more syndromes to its vocabulary, the reliance on symptom-based mapping in MEDIC is reduced. The potential problems with symptom-based mapping, while important to consider, were not determined to be of sufficient significance to deter the use of either version of the vocabulary.
Application of the extended vocabulary to MGI's annotations
With the addition of the identified missing OMIM terms and changes to the organizational structure, the extended version of MEDIC covers all mouse models of human disease currently annotated to an OMIM term in MGI. This left the set of mouse models that could not be annotated to an OMIM term. As of May 2011, there were over 250 such mouse models. Based on the existing text annotations, all of these models could be annotated to a term in the extended vocabulary. Most annotations are to general disease terms in MeSH such as Parkinson Disease (D010300) or inflammatory Bowel Diseases (D015212). A smaller set of annotations are associated to high level MeSH terms, e.g. a mouse model of congenital obstructive nephropathy (7) can be annotated to Kidney Diseases (D007674). These annotations may be useful to ontology developers to identify areas for possible term expansion.
Maintenance of the extended version of MEDIC
Ongoing curation is required to maintain the extended version of the merged vocabulary. Many of the maintenance requirements will be shared with CTD. For example, identification of changes in MeSH and OMIM, which require curatorial attention will be done using shared automated quality control processes. Modifications or additions to the OMIM to MeSH mappings for both versions of the vocabulary may be done simultaneously using a shared mapping file. The use of a shared mapping file will ensure that both versions of the vocabulary stay in sync. The actual merge process to generate the extended version and all post-merge quality control processes will need to be done at MGI. However, outputs from these quality control processes can feedback into the shared mapping file and thus improve the overall disease terminology.
As the merged vocabulary does not include all possible OMIM disease terms, ongoing curation will be required to add in existing OMIM disease terms that were not originally incorporated into the vocabulary and not identified as necessary to meet MGI's current curation needs in this review. There are ∼2200 OMIM potential disease terms that are not either in the mapping file or excluded from the mapping file for not being a disease. Not all of these terms are expected to be disease terms, some may be phenotype or enzyme activity terms (e.g. OCULAR DOMINANCE, 164 190; THEOPHYLLINE BIOTRANSFORMATION, 187 650). If a mouse model for one of the excluded diseases is identified it will be readily added to the mapping file for inclusion in the merged vocabulary. We would also recommend the creation of a tracking system, such as a SourceForge tracker, so that other groups outside of CTD and MGI may suggest additional OMIM terms to add or other changes. New OMIM disease terms are identified and incorporated as part of the current ongoing curation of MEDIC.
Current use of OMIM at MGI requires ongoing quality control and annotation updates. The most time consuming part of this work is the incorporation of updates to annotations required when OMIM refines the definition of a term. For example, in the past, OMIM changed the term PARKINSON DISEASE into the term PARKINSON DISEASE, LATE-ONSET. This required extensive annotation review and modifications of existing records to ensure annotations were consistent with this change. As well, OMIM is working to separate the phenotype and gene records (those prefixed with a + in OMIM) into individual gene (prefixed with a * in OMIM) and phenotype (prefixed with a # in OMIM) records. These changes also require modifications to MGI annotations. It is expected that adoption of the extended version of MEDIC will avoid the need to modify and update annotations, providing for a substantial curatorial time savings. For example, had the extended vocabulary been in use when OMIM changed the term PARKINSON DISEASE into the term PARKINSON DISEASE, LATE-ONSET, updates to the extended vocabulary would have been made to reflect the term change, but annotations to the MeSH term Parkinson Disease (D010300) would not have required review.
Conclusions
With the future development of a formal disease ontology uncertain, a merger of disease terms from MeSH and OMIM is a viable, practical solution to a pressing curation need. The merger of MeSH and OMIM allows access to highly detailed OMIM disease records and to the hierarchical structure and generic disease terms in MeSH. The vast majority of OMIM to MeSH mappings in the merged vocabulary are of high quality and consistency. The expanded scope and specificity of the extended version of MEDIC is able to cover all of the MGI's disease model curation needs and the process for updating and adding new mappings is quick and easy. In addition to full coverage of MGI's existing disease model annotations, access to the MeSH hierarchy allows for retrieval of disease model sets, such as all mouse models of Parkinson disease, not currently possible using OMIM alone. The use of existing vocabularies (MeSH and OMIM) makes excellent use of available resources. In addition, both versions of the vocabulary are able to inform development of more formal disease ontologies providing developers with a highly curated set of OMIM to MeSH relationships. In this vein, a file containing the extended version of MEDIC in OBO format is available from MGI (ftp://ftp.informatics.jax.org/pub/mosh) and we encourage developers of disease ontologies to use this as the basis of their MeSH to OMIM relationships. The incorporation of OMIM and MeSH identifiers into developing disease ontologies will greatly aid in adoption of the ontology by databases, such as CTD and MGI, as it will facilitate migration of existing annotations to the new ontology.
Funding
National Human Genome Research Institute/National Institutes of Health (HG000330) for the Mouse Genome Database at MGI; National Institute of Environmental Health Sciences/National Library of Medicine (ES014065); National Institute of Environmental Health Sciences (ES014065-04S1); National Center for Research Resources (P20RR016463) at CTD. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Funding for open access charge: National Human Genome Research Institute/National Institutes of Health (HG000330).
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank Drs Lynn Schriml and Warren Kibbe for their helpful discussions on the status of the Disease Ontology.