-
PDF
- Split View
-
Views
-
Cite
Cite
Terry F. Hayamizu, Sherri de Coronado, Gilberto Fragoso, Nicholas Sioutos, James A. Kadin, Martin Ringwald, The mouse–human anatomy ontology mapping project, Database, Volume 2012, 2012, bar066, https://doi.org/10.1093/database/bar066
Close - Share Icon Share
Abstract
The overall objective of the Mouse–Human Anatomy Project (MHAP) was to facilitate the mapping and harmonization of anatomical terms used for mouse and human models by Mouse Genome Informatics (MGI) and the National Cancer Institute (NCI). The anatomy resources designated for this study were the Adult Mouse Anatomy (MA) ontology and the set of anatomy concepts contained in the NCI Thesaurus (NCIt). Several methods and software tools were identified and evaluated, then used to conduct an in-depth comparative analysis of the anatomy ontologies. Matches between mouse and human anatomy terms were determined and validated, resulting in a highly curated set of mappings between the two ontologies that has been used by other resources. These mappings will enable linking of data from mouse and human. As the anatomy ontologies have been expanded and refined, the mappings have been updated accordingly. Insights are presented into the overall process of comparing and mapping between ontologies, which may prove useful for further comparative analyses and ontology mapping efforts, especially those involving anatomy ontologies. Finally, issues concerning further development of the ontologies, updates to the mapping files, and possible additional applications and significance were considered.
Database URL:http://obofoundry.org/cgi-bin/detail.cgi?id=ma2ncit
Introduction
Anatomy is an important biological integrator. Reference to anatomical structures is most often an integral component in the representation of gene expression data, biological and pathological processes, and normal and disease phenotypes. Anatomy ontologies are structured vocabularies of anatomical entities that enable the standardized description and integration of anatomical data. Numerous anatomy ontologies are being developed, including those for model organisms such as the laboratory mouse, as well as for the human. Most of these ontologies have been developed independently, with appreciable differences with regards to their scope and granularity, as well as to hierarchical organization. Many are currently being used by a variety of scientific resources to annotate a wide range of biological and biomedical data. In order to be able to integrate these data, it will be necessary to develop mechanisms with which to provide and appropriately utilize accurate cross-mappings between the various anatomy ontologies.
In order to address issues of interoperability between databases in the cancer research community, the National Cancer Institute (NCI) introduced the cancer Biomedical Informatics Grid (caBIG®) https://cabig.nci.nih.gov/ (1). One of the primary objectives of caBIG® was to enhance the dissemination of basic research results to clinical settings, and an important milestone in achieving this will require the cross-mapping of the terms and data elements as they are used in these different contexts. As part of caBIG®, the Mouse–Human Anatomy Project (MHAP) was a collaborative effort by the mouse Gene Expression Database (GXD) project (2), part of Mouse Genome Informatics (MGI) http://www.informatics.jax.org/ at The Jackson Laboratory, and the NCI Center for Bioinformatics and Information Technology (CBIIT). The objective was to facilitate the mapping and harmonization of anatomy ontologies that are currently being used for annotation of data for mouse and human models by MGI and the NCI.
As part of this study, various methodological approaches and software tools with which to perform a comparative analysis of anatomy ontologies, and to create mappings between terms within the ontologies, were identified and extensively evaluated. Subsequently, an in-depth comparison of the mouse and human anatomy ontologies were performed, and both anatomy ontologies were extended and harmonized. Appropriate matches between mouse and human anatomy terms were identified, resulting in an extensive set of mapped pairs. Links between mouse and human anatomy terms based on the mappings will facilitate closer integration of human and mouse data, promote the use of the mouse as a model for biomedical research, and accelerate translation of basic research discoveries into new clinical therapies.
Ontologies
Adult Mouse Anatomy (MA)
The mouse anatomy ontology was developed by GXD to provide standardized nomenclature for anatomical structures in the postnatal mouse (3). The MA is structured as a directed acyclic graph with multiple inheritance using both is–a and part-of relationships, and is organized in multiple ways from both spatial and anatomical system perspectives. The ontology is accessible via a browser at MGI (http://www.informatics.jax.org/searches/AMA_form.shtml), and is also available for download through the OBO Foundry (http://obofoundry.org/). Currently, containing approximately 3000 unique terms, the MA continues to be expanded and refined in response to additional sources of information, and according to the needs of the scientific community. MA terms and identifiers are now being used by a number of database resources in descriptions of gene expression patterns and other biological data pertinent to mouse anatomy, including GXD, Pathbase (4), the Mammalian Phenotype (MP) Ontology (5) and for the annotation of mouse gene products using the Gene Ontology (GO) (6).
NCI Thesaurus (NCIt)
The NCI Thesaurus is a large reference terminology and biomedical ontology developed by the NCI as part of Enterprise Vocabulary Services (EVS) http://www.cancer.gov/cancertopics/cancerlibrary/terminologyresources, and is used for data systems by the NCI and others. NCIt provides structured representation of over 90 000 cancer-related concepts for basic and translational research, as well as for clinical care (7,8). The ontology is structured as a subsumption hierarchy with additional relationships providing logical links between concepts. The NCIt can be accessed using a web browser (http://ncit.nci.nih.gov/ncitbrowser/) and is also available for download in several file formats from that website.
Background and previous work
An initial evaluation involved a preliminary comparison of the MA ontology and NCIt, primarily to determine the feasibility of the proposed mapping between terms. One of the goals of this work was to provide an estimate of the number of concepts in the existing ontologies that could be mapped directly. Several different approaches were utilized to identify matching concepts within the two ontologies, including a simple lexical comparison of MA terms with the full NCI Thesaurus, and a preliminary manual comparison of the MA terms against a list of NCIt (human) anatomy terms. In addition, a group at the National Library of Medicine analyzed the ontologies using a combination of lexical and structural similarity methods (9). The results of each approach were then re-verified by manually curated analysis, which involved side-by-side comparison of terms within the ontologies using the web-based ontology browsers provided by MGI and the NCI.
With regards to the different approaches to matching terms from the two ontologies, the automated methods were much faster, identified some valid matches that had been missed by the manual evaluation and also pointed out some errors in the manual mapping process. However, there were more false negative (16.0% versus 7.32%) and slightly more false-positive results (2.1% versus 1.15%) with the automated approach. Manual evaluation and re-evaluation of the results of either method by a ‘domain expert’, although more labor intensive, was absolutely critical for validation. Furthermore, each of the methods picked up a significant number of valid matches that had been missed by the other approach. Detailed results from comparison of the lexical and structural similarity approach with manual curation, including specific examples, were reported in a previous publication (10).
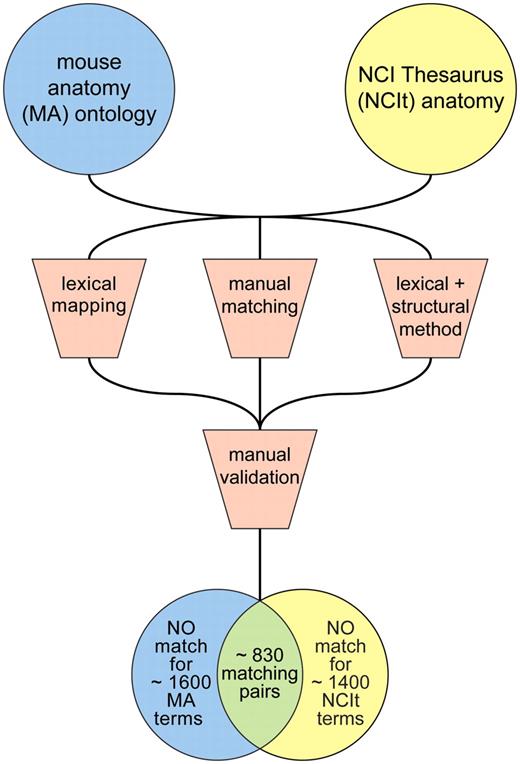
Based on this work, we estimated that valid matches could be made for approximately one-third of the existing MA and NCIt anatomy terms (Figure 1). A majority of the matches were identified by each of the approaches used, providing further support for their validity. Significant progress was also made toward identifying non-matching concepts, which was particularly informative in terms of recognizing the types of terms that were represented in one of the ontologies but not in the other. Broad identification of sets of terms that were not shared between the independent ontologies, as well as those that were shared, was useful in planning for subsequent steps in the process.
Preliminary identification of equivalent terms using a combination of automated and manual curation approaches.
Evaluation of tools
Much of the preliminary manual analysis of the MA and NCIt terms was carried out using web-based browsers for the respective ontologies at MGI and NCI. For a more comprehensive and thorough comparison and mapping of terms between the anatomy ontologies, however, it was apparent that more sophisticated tools would be required. Several potentially useful ontology building and mapping applications were identified. Of those, the following tools were chosen for further evaluation: DAG/OBO-Edit (11), Protégé-OWL (12) and COBrA (13). The specific versions used for the initial evaluation of ontology editing tools were: DAG-Edit 1.418, OBO-Edit 1.001, COBrA 1.0 and Protégé 3.1 beta (build 185), with the Prompt plug-in (for Protégé).
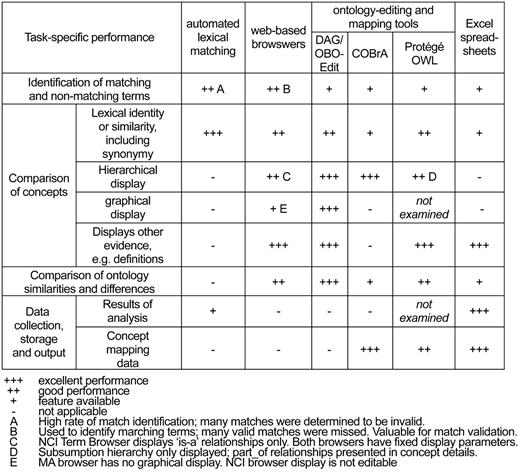
Owing to somewhat different intended uses for these tools and applications, it was apparent that each would have different strengths and limitations. We elected to focus our performance evaluation on the set of specific activities required for the MHAP: (i) Identification and validation of potentially matching anatomical terms, with regards to lexical, structural, definitional and other criteria; (ii) Comparison of similarities and differences between the ontologies, including overall ontology structure and levels of granularity; (iii) Actual mapping of concepts to one another, with results available in output format(s) appropriate to the requirements of potential users; and (iv) Collection and storage of data from the analysis, in format(s) amenable to future analysis. Each of the methods and tools specified for this work were tested for each of these activities. Results from this analysis are summarized in Figure 2.
Task-specific performance features at the time of the tools evaluation. Since that time, both OBO-Edit and Protégé have added functionality in the form of new features as well as with plug-ins.
Overall, it was determined that many factors can influence the overall performance of the different analytic methods and tools, and distinct features of each may impact its utility with regards to task-specific performance. Consequently, selection of the optimal methods and/or tools would be highly dependent on the precise nature of the analysis being proposed. The ability to thoroughly review ontology terms, including synonyms and definitions (when available), as well as their hierarchical context within the ontologies, would be critical to this effort. In this regard, each web-based browser and ontology-editing tool provided a different set of concept information, as well as different view options. All were useful in these efforts. Notably, the graphical display features provided by some browsers and editing tools enabled different views of concepts within their respective ontology and were, thus, extremely useful in much of the analyses.
An important finding was that none of the methods and/or tools evaluated were able to provide the entire range of features required to best perform all of the tasks proposed for this project. Specifically, some tools were better for comparing terms within the ontologies in a comprehensive way, whereas others were better able to provide mappings between terms. Among the additional features that would have provided significant utility was a means with which to view multiple ontologies and specific concept information simultaneously side-by-side, in hierarchical formats showing all relevant relationship types, as well as in an editable graphical format. Furthermore, none of the originally proposed tools provided an adequate way of collecting, storing and organizing the data collected from the various types of comparative analyses. In this regard, spreadsheets were found to be essential for such ‘mundane’ tasks as sorting and grouping sets of terms, as well as for providing a comprehensive record of the details of the analysis. The spreadsheet format was also useful in creating customizable reports of various aspects of the analysis as well as for specifically exporting the mapping results (see below).
It should be noted that, since the time of our evaluation, numerous additional tools have been developed, and software providing a wide range of added functionality is now available for the applications used in this effort (e.g. in the form of OBO-Edit and Protégé plug-ins). With ontology mapping efforts becoming more prevalent, we clearly anticipate that software developers will continue to address the kinds of issues we have encountered. Significant improvements in automated ontology alignment methods are expected as well. However, we envision that a combination of methodological curatorial approaches and software tools will continue to be required for the range of different tasks involved in these types of efforts.
Comparative analysis of the ontologies
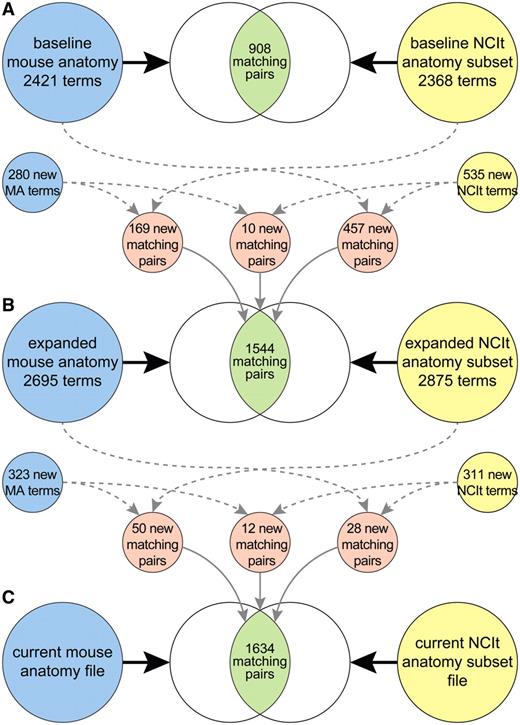
Using the tools selected in the previous task, an in-depth comparative analysis of the existing mouse and human anatomy ontologies was performed. For this task, a set of MA terms (file date 28 March 2005) was compared with human anatomy concepts in the Anatomic_Structure_System_or_Substance branch of the NCIt (version 05.03d). This subset excluded the NCIt sub-branches of Cell_Part, Cell_Structure, Extracellular_Space, Gene_Physical_Location, Macromolecular_Structure, Normal_Cell and Embryological_Structure_or_System since these domains are not represented in the MA. The analysis involved examination of each of the matched term pairs that had been identified by various approaches in the preliminary work, and then manually validating the matching. In most cases, validation of matched pairs was straightforward and easily identifiable by any appropriate ‘domain expert’. In the remaining cases, further validation consisted of a comprehensive analysis of all available evidence provided by the MGI and NCIt resources, including synonymy and definitions (when available), as well as the structural context for the terms within the ontologies. Overall, a total of 908 validated matches were identified between MA and NCIt human anatomy terms during this phase of the study (Figure 3A).
Mouse and human anatomy ontology terms, and corresponding matched sets of terms, at start of study (A), after extension and harmonization of the ontologies (B) and after recent updates to the files (C).
Throughout this analysis, it was apparent that the basic structural organization as well as the overall content of the anatomy ontologies were more similar than different. In general, differences between the anatomy ontologies reflected differences specifically with regards to the following factors: (i) hierarchical organization; (ii) ontology coverage; and (iii) granularity.
- (i)
Subsumption hierarchy requirements, accommodation of tissue sampling issues and other factors resulted in different choices for root nodes and other high-level concepts. For instance, the MA had primarily focused on partonomic anatomical relationships and did not include terms corresponding to the high level NCIt classes of Body_Part, Organ or Tissue. Thus, subclasses of these terms were represented somewhat differently in the MA than in the NCI Thesaurus. For example, the MA term central nervous system is a part_of nervous system, whereas the equivalent NCIt concept is subclass of Nervous_System_Part, as well as part of Nervous_System.
Furthermore, a number of MA terms had been included to accommodate different views of the hierarchy, as well as tissue sampling issues found to be important for data collection. For instance, in order to accommodate the need to represent certain tissues within specific anatomical regions, ‘group’ terms (e.g. head skin and limb muscle) have been included as subterms, when appropriate, as part of those regions. Some of these distinctions are conceptual and by their nature may be somewhat arbitrary. However, different breakdowns of the anatomy were considered to be required in order to appropriately annotate, for example, different types of expression and phenotype data.
- (ii)
Many specifically named anatomical structures were represented in only one of the ontologies. For example, a number of these are specifically named arteries, veins and muscles. Representation of these structures in the MA is particularly extensive since they are major components of several of the primary MA resources used in developing the ontology. Since a majority of these structures are likely to be present in both mouse and human, these terms were considered as candidates for adding to the other ontology.
- (iii)
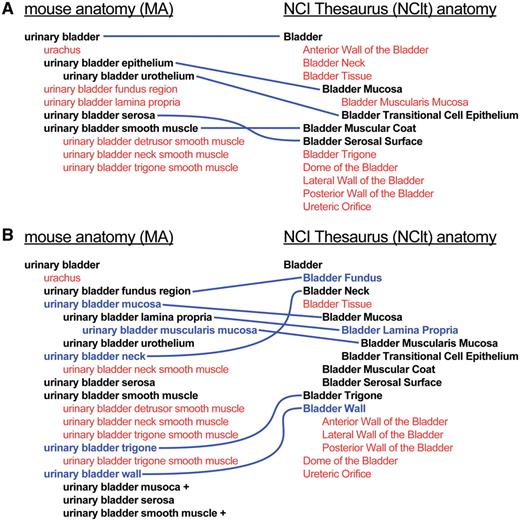
Much of the dissimilarity observed between the anatomy ontologies reflected differences in the level of detail in which substructures were represented in each ontology. An example of this is shown in Figure 4A. In this case, the MA has focused primarily on tissue components of the urinary bladder, whereas the NCIt has a more detailed representation of regional parts. In another case, MA parts of foot included tarsus, metatarsus and foot digit, the latter with subconcepts including foot digits 1, 2, 3, 4 and 5, whereas the NCIt had Dorsal_Region_of_Foot, Plantar_Region and Toe.
Example of the extension and harmonization process. (A) Representation of the urinary bladder in the MA (left) and NCIt (right) prior to the revision process. Terms in black, linked by blue lines, represent matched sets of terms; terms in red are those not shared by the other ontology. (B) Urinary bladder concept subtrees after extension and harmonization of the ontologies. Terms in blue are those that have been added, with blue lines indicating corresponding terms in the other ontology.
In summary, comparison of the MA with the NCI Thesaurus anatomy subsection identified many terms specific to one of the anatomy ontologies, but a very limited number of these represented actual species-specific anatomical concepts. Mouse-specific anatomical structures included the tail and its substructures, muzzle/snout, coat hair and vibrissa. NCIt terms without true mouse equivalents included Eyebrow and Sacrum, as well as those reflecting differences in representation and coverage of the Breast and Prostate. Most differences appeared, instead, to be a consequence of decisions made with regard to the scope of each ontology rather than actual differences between the organisms themselves. Thus, while it was determined that all terms would not, and in fact should not, be mapped to the other ontology, it was also apparent that both anatomy ontologies would derive significant benefit from extensions and other modifications resulting from the harmonization effort.
Extending and harmonizing the ontologies
The first step toward harmonizing the existing adult mouse and human anatomy ontologies was to develop specific plans, both for extending and for harmonizing the ontologies. Specific guidelines were established prior to initiating the effort: (i) Addition of terms was considered in situations where a concept was represented in one ontology but not in the other; (ii) Term names and hierarchical organization were modified when feasible to facilitate harmonization between the ontologies; (iii) Vocabularies were augmented with synonymy to accommodate different naming conventions; and (iv) Addition of specific classes previously not included in the domain of an ontology was carefully considered. Other changes requiring more substantial modifications to the existing ontologies were identified for consideration but, for the most part, deemed to be beyond the scope of this specific project. Subsequently, changes were made to both the MA ontology and NCIt human anatomy subsection, including creating additional terms and modifying existing terms and hierarchies where appropriate. An example of changes resulting from the extension and harmonization effort is shown in Figure 4B.
In summary, the baseline MA file contained 2421 terms, from which 5 pairs of terms were subsequently merged and 1 term was deleted. As a result of changes made to the MA, including those directly related to the extension and harmonization effort, 280 new terms were added. This resulted in an updated MA file (dated 20 January 2006) with 2695 terms and 169 additional matches with NCIt terms. Similarly, the NCIt anatomy subset that served as the baseline included 2368 terms. Concurrent with additions to the MA, changes to the NCIt resulted in an updated human anatomy file (based on version 06.01c) containing 2875 terms. Specifically, 535 terms were added to the NCIt anatomy subset. Of these, 457 matched existing MA terms, whereas 10 new mappings resulted between terms that were new to both the MA and NCIt. Pursuant to extension and harmonization of the anatomy ontologies, 636 additional matches were identified, resulting in a total of 1544 valid matches (Figure 3B).
Our analysis also revealed several cases of ‘redundant’ mappings in which a given anatomy term potentially matched more than one term in the other ontology. Some of these identified situations in which two terms within an ontology represented the same anatomical entity and, thus, were candidates for merging or for retirement of one of the terms. Other cases were less straightforward and, in some situations, would likely require considerable ontology revision, as well as data re-annotation, in order to resolve the issue. Given the limited scope of the MHAP project, the decision was made to allow for multiple mappings in cases where the matches, although not strictly equivalent, might nonetheless provide appropriate and valuable links between mouse and human data. For example, the MA term ‘mammary gland’ (with the synonym ‘breast’) was mapped to both the NCIt term ‘Mammary Gland’ and to the widely used human-specific term ‘Breast’. While ‘Mammary Gland’ and ‘Breast’ are not strictly equivalent, much of the data annotated to ‘mammary gland’ in the mouse would, in fact, be annotated to ‘Breast’ and not to ‘Mammary Gland’ for the human.
Recent updates to the mapping file
During the project, substantial changes were made to both the MA and the NCIt anatomy subsection to optimize harmonization of the anatomy ontologies, and additional valid matches between corresponding mouse and human terms were subsequently identified. The table of mouse–human mappings needed to be updated accordingly. Thus, it was recognized from the onset that, since both ontologies would be continually refined and expanded based on additional resources and community needs, the process of identifying and validating additional mouse–human matches will also need to be periodically reiterated to maintain accurate mapping between the mouse and human anatomy terms.
Subsequent to the extension and harmonization phase of this study, 323 new terms were added to the MA (based on file dated 15 July 2011). Concurrently, 311 relevant terms (i.e. within the domain of the MA) have been added to the NCIt (version 11.09d). When each of the added terms was analyzed with regards to possible corresponding terms in the other ontology, it was determined that 50 of the new MA terms could be mapped to existing NCIt terms, whereas 28 of the new NCIt terms could be mapped to existing MA terms. In addition, 12 mappings could be made between new MA and new NCIt terms. Thus, 90 additional mappings were identified for a revised total of 1634 matched sets of terms in the updated mapping file (Figure 3C).
Mappings between mouse and human anatomy terms
An important product from this work was the identification of matches between adult mouse and human anatomy terms, which could be used to facilitate cross-linking between data resources using the anatomy ontologies. Information regarding validated mappings, including term names and numerical identifiers, was collected and stored throughout the project in spreadsheets. The spreadsheet data could be readily transformed into a variety of output formats, including tables and tab-delimited text files. An interim version of the mappings file has previously been made available upon request, and was also included in a collection of mapping sets for various biomedical ontologies at the BioPortal website. The updated mappings have now been made available for download as an obo-formatted file through the OBO Foundry: http://obofoundry.org/cgi-bin/detail.cgi?id=ma2ncit. Pursuant to ongoing development of both MA and NCIt anatomy ontologies, the mouse–human anatomy mappings will continue to be revised and the OBO Foundry file updated accordingly.
Interaction with other mapping efforts
Ontology alignment evaluation initiative
The ontology alignment evaluation initiative (OAEI) (http://oaei.ontologymatching.org/) is a collaborative effort in the ontology alignment community aimed at rigorous and extensive evaluation of ontology alignment technologies (14). Since 2007, the OAEI has used the mouse–human anatomy set, with some modifications, as a ‘gold standard mapping’ example of a ‘real world case’ in an annual competitive evaluation of ontology matching approaches. Feedback from the OAEI has also led to updates to the mappings file.
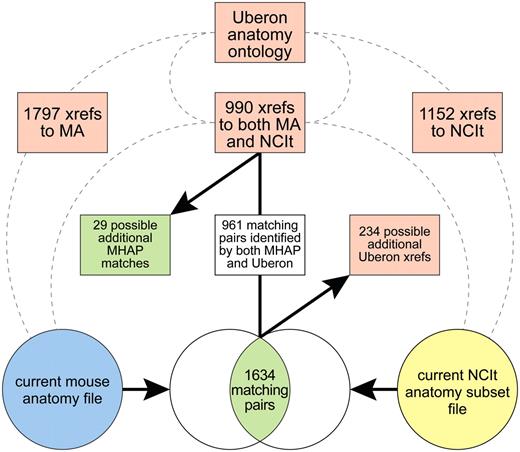
Uberon
Uberon (http://obofoundry.org/wiki/index.php/UBERON:Main_Page) is an integrated cross-species anatomy ontology constructed using a combination of semi-automated methods and manual curation (15). The ontology consists of classes representing anatomical entities that are shared across a variety of metazoan organisms. The Uberon file contains extensive cross-references between its terms and other anatomy ontologies, including the MA and NCIt, which are maintained as semantic-free ‘xref's.
Using an Uberon file (data version 2011-08-04) downloaded from the Open Biological and Biomedical Ontologies (OBO) website (http://obofoundry.org/), we found 1797 Uberon terms with xrefs to the MA, 1152 with xrefs to NCIt and 990 with xrefs to both MA and NCIt (Figure 5). When compared with the updated set of MHAP mappings, 961 of the 1634 were also identified by Uberon xrefs to both ontologies. Of particular interest, in 29 cases, an Uberon term had xrefs to both the MA and NCI, but these had not yet been identified as MHAP mappings. These will be further evaluated and, if validated, used to update the mouse–human anatomy mappings. In addition, for 231 of the terms mapped by the MHAP, Uberon had xrefs to only the MA term, whereas three mappings had an Uberon xref to only the NCIt term, indicating that the MHAP mappings may be a resource for potential additional xrefs for Uberon.
Summary and conclusions
During the course of this project, methodological approaches and software tools with which to perform a comparative analysis of anatomy ontologies and to create mappings between terms within the ontologies were identified and evaluated. It was determined that distinct features of the individual tools impact their utility with regards to task-specific performance, and that separate tools, combinations thereof, or additional tools, would likely be required for any endeavor of this kind.
Automated methods and manual curation were utilized to carry out a comprehensive comparative evaluation of the MA and the NCIt human anatomy ontologies, which included a detailed analysis of similarities and differences between them. Manual curation was found to be critical in this regard. Subsequently, the anatomy ontologies were extended and harmonized, and appropriate matches between mouse and human anatomy terms were identified. Ongoing efforts include continued development of the MA and NCIt anatomy ontologies, with plans for periodic updates of the mouse–human anatomy mappings file, which will continue to be made available.
The laboratory mouse serves as a premier animal model for biomedical research. Terms from the MA ontology are currently being used by a number of database resources to describe and integrate biological information about the mouse pertinent to anatomy such as gene expression, biological and pathological processes, and phenotype data. Likewise, the anatomical concepts in the NCI Thesaurus are and will be used in similar ways to record and integrate different types of cancer-related human data within the caBIG® framework. Thus, cross-mappings between the anatomical ontologies will facilitate the integration of mouse and human data, and promote the translation of basic research discoveries into clinical settings.
Funding
National Cancer Institute at the National Institute of Health, Cancer Biomedical Informatics Grid (caBIG®) (project number caBIG-VCDE-14-02-02); and National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant number HD062499). Funding for open access charge: NIH (grant HD062499).
Conflict of interest. None declared.
Acknowledgements
We thank Connie Coon and Frank Hartel for contributions to the NCIt; Olivier Bodenreider and Songmao Zhang for their work aligning the ontologies using lexical and structural similarity approaches; Elena Beisswanger and the OAEI for helpful suggestions with regards to updating the mappings; Chris Mungall for assistance in making the mappings available through the OBO Foundry and, with his colleagues, for work on the Uberon ontology; Brian Davis and the Vocabularies and Common Data Elements (VCDE) Workspace group for support and assistance with caBIG® tasks; and our MGI and NCI colleagues for their advice and support.
References
Author notes
Present address: Nicholas Sioutos, Lockheed Martin Corporation, Fairfax, VA 22031, USA