-
PDF
- Split View
-
Views
-
Cite
Cite
Aurélie Névéol, W. John Wilbur, Zhiyong Lu, Improving links between literature and biological data with text mining: a case study with GEO, PDB and MEDLINE, Database, Volume 2012, 2012, bas026, https://doi.org/10.1093/database/bas026
Close - Share Icon Share
Abstract
High-throughput experiments and bioinformatics techniques are creating an exploding volume of data that are becoming overwhelming to keep track of for biologists and researchers who need to access, analyze and process existing data. Much of the available data are being deposited in specialized databases, such as the Gene Expression Omnibus (GEO) for microarrays or the Protein Data Bank (PDB) for protein structures and coordinates. Data sets are also being described by their authors in publications archived in literature databases such as MEDLINE and PubMed Central. Currently, the curation of links between biological databases and the literature mainly relies on manual labour, which makes it a time-consuming and daunting task. Herein, we analysed the current state of link curation between GEO, PDB and MEDLINE. We found that the link curation is heterogeneous depending on the sources and databases involved, and that overlap between sources is low, <50% for PDB and GEO. Furthermore, we showed that text-mining tools can automatically provide valuable evidence to help curators broaden the scope of articles and database entries that they review. As a result, we made recommendations to improve the coverage of curated links, as well as the consistency of information available from different databases while maintaining high-quality curation.
Database URLs:http://www.ncbi.nlm.nih.gov/PubMed, http://www.ncbi.nlm.nih.gov/geo/, http://www.rcsb.org/pdb/
Introduction
Background
High-throughput experiments and bioinformatics techniques are creating an exploding volume of data that are becoming overwhelming to keep track of for biologists and researchers who need to access, analyze and process existing data. Much of the available data are being deposited in specialized databases such as the Gene Expression Omnibus (GEO) (1) for microarrays or the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) (2) for protein structures and coordinates. Data sets are also being described by their authors in publications archived in literature databases such as MEDLINE and PubMed Central (PMC). Therefore, for complete information about a data set, it is important that users can easily access relevant publications from biological databases, and conversely, access the relevant biological database entry from a publication describing a specific data set. The curation of such links between data sets and publications is currently performed manually by the curators of the databases involved (biological or literature databases), who act independently based on their own criteria and methods for recording and displaying the links. In some databases (e.g. PDB), link curation is automatically generated based on the information supplied by the authors at the time of data set submission. Journal editors have recognized the importance of data sharing and of promoting the availability of links between data sets and publications by instigating editorial policies requiring authors to deposit data sets described in an article (3) and to provide specific information regarding the deposition in the article. Although these guidelines are not always strictly followed (4), data set deposition and subsequent deposition declarations in research articles are becoming common. As a result, many full-text articles [rather than abstracts (5)] contain deposition statements, that is, statements that report the deposition of a data set into a biological database by the authors. Pending validation by the curators, author reports of data deposition are the primary method for identifying links between data sets and related literature. When submitting a data set to a database, authors are asked to provide the reference to a research article describing the creation of the data set; however, if the article is not published at the time of data set submission, the authors may not be able to supply this information. Similarly, when reporting on a data set in a research article, authors are required to submit the data set to a database and to supply the corresponding accession numbers in the article; however, the full accession details may not be available at the time of the article acceptance so that the authors are sometimes unable to supply this information. For these reasons, discrepancies may arise in the information recorded in biological databases versus the literature. The objective of this article is to perform a systematic study of these discrepancies and to assess the use of automatic methods to assist curators in maintaining high-quality curation and in bridging gaps between information curated in multiple sources. To this end, we cross-examine curated links available from multiple sources and rely on full-text analysis to automatically extract evidence statements supporting the link curation suggested by each source. As a starting point, we have focused our study on two specific databases curating microarray and protein-related data: GEO and PDB. There are two main contributions of this study: first, it provides a comprehensive analysis of the existing sources of links between two biological databases and the literature, including three sources of curated links and one source of automatically extracted links. To the best of our knowledge, this is the first study comparing the curation of links between biological data sets and research articles in biological versus literature databases. Second, this study shows how a text-mining tool can help bridge the gap between curated sources and yield recommendations to improve link curation.
Related work
Applications of text-mining to complex database curation tasks
In the past decades, much research in natural language processing and text-mining for the biomedical domain has focused on named entity recognition [e.g. (6,7)], concept identification (8,9) and controlled vocabulary indexing [e.g. (10,11)]. Good performance can now be obtained for these tasks, which paves the way for efforts geared toward more complex tasks (12), including the extraction of relationships between entities, concepts and databases. Toward these goals, the Semantic MEDLINE project goes beyond keyword queries to enable MEDLINE searches based on relationships between concepts (13). The recent BioCreative III Workshop (14) comprised a ‘Gene Normalization’ task (15) that challenged participants to automatically extract links between gene mentions in the text of articles from the literature and records from the Entrez Gene database. Other recent work focused on specific data types, such as brain regions (16) and DNA sequences (17), to automatically extract links between articles in the literature and relevant biological databases–namely, five neuroanatomical databases (16) and the sequence database Genbank (17). The application goal of such research projects is to provide practical tools that database curators can use to help them in their daily routine. Although contributing to the day-to-day development and maintenance of large curated databases has become a reality for some tools (11–18), other efforts are focusing on providing evidence for a posteriori validation and improvement of curated data (19–20).
Link extraction based on full-text processing
It is important to point out that much recent work addressing link extraction and other complex data curation tasks has been possible because of the increasing availability of full-text articles versus abstracts only. Prominent sources of full-text articles in the scientific literature are the Public Library of Science (PLoS) (21) and PMC and its subset PMC Open Access (http://www.ncbi.nlm.nih.gov/pmc/about/openftlist.html). Other projects have made smaller-scale full-text corpora available (22). Based on PMC, Cohen et al. showed that the content of abstract text and full-text was significantly different (23). Although there is little benefit from full-text processing for some applications such as MeSH indexing (24), it is crucial to other tasks such as the extraction of scientific claims: a recent study showed that only 7.84% of scientific claims are reported in abstracts (25). Similarly, data deposition is mentioned much more frequently in full-text than it is in abstract text: about 6% of all full-text articles in PMC contain a deposition statement versus <0.01% of abstracts (5). Some of the earliest work addressing the creation of links between biological databases and the literature based on full-text analysis relied on selected PMC full-text XML files. It provided an enhanced version of full-text articles by integrating clickable links to PDB and Gene Ontology within the full-text of articles accessible from the BioLit portal (26). It also included clickable links to PDB entries from occurrences of the accession codes within the full-text. However, no distinction is made between newly deposited data, reused data or commented data. For this reason, this work can be described as browsing-oriented rather than curation-oriented.
Material and methods
Biological and literature databases
In previous work (5), we characterized data deposition in biological databases through an analysis of data deposition statements in PMC articles. See online supplementary material that presents the distribution of databases mentioned in a random sample of deposition sentences automatically extracted. It can be seen that GenBank, GEO and PDB are the most prevalent databases in our data set. They are major databases receiving data deposition, but it could also reflect the bias of our training set for the tool. In this study, we focused on the two biological databases where researchers routinely deposit microarray and protein-related data, namely GEO and PDB, respectively. Our goal was to survey the links available between these databases and the literature available in PubMed.
The GEO (http://www.ncbi.nlm.nih.gov/geo/) is a public functional genomics data repository supporting MIAME-compliant data submissions (Minimum Information About a Microarray Experiment). Array- and sequence-based data are accepted. Tools are provided to help users query and download experiments and curated gene expression profiles. Table 1 shows a description of the various data types stored, along with the number of entries of each type available from the repository. At the time of submission, authors may supply the reference to a journal article reporting on the creation of the data set so that a link to the publication can be included in the GEO entry. GEO curators check and supplement publication information submitted by the authors as needed.
List and description of data types available from GEO
| Data type . | Characteristics . | Accession . | Number . |
|---|---|---|---|
| Platforms | A platform may refer to many samples that have been submitted by multiple submitters. | GPL | 9354 |
| Samples | A sample entity must refer to only one platform and may be included in multiple series. | GSM | 6 31 997 |
| Series | A series record links together a group of related samples and provides a focal point and description of the whole study. | GSE | 25 447 |
| Data sets and profiles | Selected primary records undergo an upper-level of rendering into Data set and gene profile records. | GDS | 2720 |
| Data type . | Characteristics . | Accession . | Number . |
|---|---|---|---|
| Platforms | A platform may refer to many samples that have been submitted by multiple submitters. | GPL | 9354 |
| Samples | A sample entity must refer to only one platform and may be included in multiple series. | GSM | 6 31 997 |
| Series | A series record links together a group of related samples and provides a focal point and description of the whole study. | GSE | 25 447 |
| Data sets and profiles | Selected primary records undergo an upper-level of rendering into Data set and gene profile records. | GDS | 2720 |
List and description of data types available from GEO
| Data type . | Characteristics . | Accession . | Number . |
|---|---|---|---|
| Platforms | A platform may refer to many samples that have been submitted by multiple submitters. | GPL | 9354 |
| Samples | A sample entity must refer to only one platform and may be included in multiple series. | GSM | 6 31 997 |
| Series | A series record links together a group of related samples and provides a focal point and description of the whole study. | GSE | 25 447 |
| Data sets and profiles | Selected primary records undergo an upper-level of rendering into Data set and gene profile records. | GDS | 2720 |
| Data type . | Characteristics . | Accession . | Number . |
|---|---|---|---|
| Platforms | A platform may refer to many samples that have been submitted by multiple submitters. | GPL | 9354 |
| Samples | A sample entity must refer to only one platform and may be included in multiple series. | GSM | 6 31 997 |
| Series | A series record links together a group of related samples and provides a focal point and description of the whole study. | GSE | 25 447 |
| Data sets and profiles | Selected primary records undergo an upper-level of rendering into Data set and gene profile records. | GDS | 2720 |
The PDB archive (http://www.rcsb.org/pdb/) contains information about experimentally determined structures of proteins, nucleic acids and complex assemblies. When submitting a data set described in an article, the PDB policies recommend that authors and/or journals notify the curators so that a link between the data set and publication can be recorded in the entry.
MEDLINE (http://www.ncbi.nlm.nih.gov/PubMed) is the reference database for citations of published literature in the biomedical domain. As of November 2011, it contained >19 million citations. In 1988, MEDLINE curators started to record information pertaining to the registration of several types of biological data in the Secondary Source ID (SI) field of citations. The first type of biological data to be processed in this way was the sequence data deposited in GenBank and discussed in articles cited in MEDLINE. Specific technical and historical details about SI indexing can be found on the National Library of Medicine (NLM) website at http://www.nlm.nih.gov/bsd/mms/medlineelements.html#si. As molecular sequence databases became increasingly prevalent and used by researchers, additional databases were included in the curation workflow over the years. For instance, PDB (27) was added in 1993 and GEO in 2006 (28). Currently, 13 databases are being linked to MEDLINE articles through (SI) curation, including clinical trials (using identification numbers obtained through: http://isrctn.org/) and PubChem substances (http://pubchem.ncbi.nlm.nih.gov/). In the case of GEO, it can be noted that the four data types listed in Table 1 are unevenly curated in the (SI) field. As shown in Table 2, the GEO data type that is most frequently curated in MEDLINE is ‘Series’ with 3208 citations corresponding to 3883 GEO records (more than one record may be reported in a single article).
Number of MEDLINE citations with curated GEO data
| Data type . | Accession . | Citations . | GEO records . |
|---|---|---|---|
| Platforms | GPL | 94 | 105 |
| Samples | GSM | 220 | 1680 |
| Series | GSE | 3208 | 3883 |
| Data sets and profiles | GDS | 7 | 7 |
| Data type . | Accession . | Citations . | GEO records . |
|---|---|---|---|
| Platforms | GPL | 94 | 105 |
| Samples | GSM | 220 | 1680 |
| Series | GSE | 3208 | 3883 |
| Data sets and profiles | GDS | 7 | 7 |
Number of MEDLINE citations with curated GEO data
| Data type . | Accession . | Citations . | GEO records . |
|---|---|---|---|
| Platforms | GPL | 94 | 105 |
| Samples | GSM | 220 | 1680 |
| Series | GSE | 3208 | 3883 |
| Data sets and profiles | GDS | 7 | 7 |
| Data type . | Accession . | Citations . | GEO records . |
|---|---|---|---|
| Platforms | GPL | 94 | 105 |
| Samples | GSM | 220 | 1680 |
| Series | GSE | 3208 | 3883 |
| Data sets and profiles | GDS | 7 | 7 |
Sources of link curation and analysis of the existing links
For GEO, based on the observation that series records were the most prevalent in MEDLINE curation (see Table 2), we decided to focus the study on this data type only. On 1 November 2011, we downloaded 25 715 GEO Series Simple Omnibus Format in Text (SOFT) files from the GEO website (ftp://ftp.ncbi.nih.gov/pub/geo/) and extracted the links between the series accession number and PubMed Identifier (PMID) when available. For example, the sample data shown in Figure 1 resulted in the extracted link between PMID 21772264 and GEO ID GSE26151.
For PDB, on 1 November 2011, we also downloaded the ‘Primary Citation’ report for the 76 288 structures available in PDB at that time. The data were available as a comma-separated values (csv) file from which the information of interest could be directly extracted: PDB accession number and relevant PMID.
For MEDLINE, on 1 November 2011, we downloaded the citations that had a GEO or PDB accession number using simple PubMed queries ‘GEO (SI)’ and ‘PDB (SI)’. The links between PMID and accession numbers were extracted from the relevant fields of the MEDLINE format citation. For example, the sample data shown in Figure 2 resulted in the extracted link between PMID 16436444 and GEO ID GSE3028. Links relevant to series records only were selected for the study. However, for simplicity, in the remainder of this article we refer to these links as the GEO links.
Additionally, we also consider the results of an automatic link curation tool described later in the text. Through a simple comparison of the link sources, our goal is to determine how exhaustive the pool of curated links is, in addition to assessing the overlap between sources.
Text-mining tool for supporting link identification
In recent work (5), we developed a tool that automatically processes full-text articles to determine whether the article is relevant for link curation, that is, whether the article can be considered as the primary citation for the biological data it describes. The tool also automatically extracts statements supporting the classification decision. For example, for PMID 21282644, the tool predicts that the article is relevant for link curation and extracts the following statement as supporting evidence: ‘Data deposition: the data reported in this article has been deposited in the GeoArchive database (GEO accession no GSE2350 and GSE26408).’
In this work, we use this tool to automatically retrieve evidence statements from articles with conflicting curation status from the different sources available to us: MEDLINE, the biological databases (GEO and PDB) and the automatic relevance prediction from the tool.
Evaluation of automatically extracted evidence statements
In spite of the good performance of the tool described previously in the text [81% F-measure, as reported in (5)], we are aware that the tool’s results may be erroneous. For a more specific assessment of the tool’s value as an aid to curators in the case of GEO and PDB links to the literature, we manually assessed the evidence statements automatically retrieved by the tool by classifying them into four main categories:
Deposition: If the evidence statement shows the data were deposited.
19706781|The microarray data have been submitted to GEO (http://www.ncbi.nlm.gov/geo/) under the accession GSE13451.
Reuse: If the evidence statement shows that the data were reused and not deposited.
20673354|We downloaded the normalized data of four breast cancer gene expression data sets from GEO http://www.ncbi.nlm.nih.gov/geo/ [23-26]
Ambiguous: If the evidence statement is not clear-cut about the deposition of data. This is often the case with incomplete or inconclusive statements.
21410990|cel files from the NCBI GEO database (accession number: GSE11045) and raw NGS
20805289|Data used in this analysis are publicly available at NCBI's GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession series numbers GSE11624, GSE7448 and GSE16374.
Comment: If the evidence statement is providing comments about the database or data available in the database.
17993534|This finding was confirmed by primer extension analysis (data not shown) and by recently deposited microarray data in the GEO database (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE8478)
Results
Analysis of existing link sources
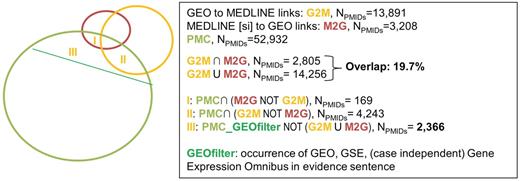
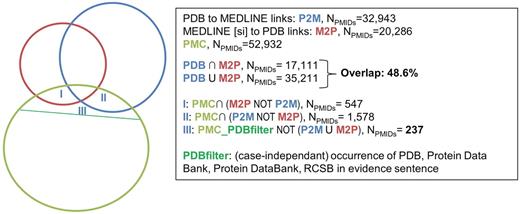
Figure 3 shows the overlap between GEO (in orange) and GEO curation in MEDLINE (SI) (in red), whereas Figure 4 shows the overlap between PDB (in blue) and PDB curation in MEDLINE (SI) (in red). In terms of impacted articles, the overlap is low for both sources: 19.7% for GEO and 48.6% for PDB. This means that more than half of the articles describing data deposited in one of the biological databases are not curated either in the literature or in the database itself. It can be noted that in both cases, the number of curated articles is higher in the biological database versus literature database (13 891 curated articles in GEO vs. 3208 for GEO_MEDLINE; 32 943 curated articles in PDB vs. 20 286 in PDB_MEDLINE).
Overlap between GEO, MEDLINE (SI) and the results of text-mining on PMC; evidence statements were extracted from full-text articles for three categories that were outside the consensus between link sources: (I) Articles curated in MEDLINE but not by GEO, (II) articles curated by GEO but not by MEDLINE and (III) articles curated neither by MEDLINE nor GEO, but identified as relevant for link curation for GEO by our automatic tool.
Overlap between PDB, MEDLINE (SI) and the results of text-mining on PMC; evidence statements were extracted from full-text articles for three categories that were outside the consensus between link sources: (I) Articles curated in MEDLINE but not by PDB, (II) articles curated by PDB but not by MEDLINE and (III) rticles curated neither by MEDLINE nor PDB, but identified as relevant for link curation for PDB by our automatic tool.
The figures also display the number of full-text articles from PMC that were automatically identified as relevant for link curation (in green). Although the green set of PMC articles automatically found relevant for link curation is the same in Figures 3 and 4, filters are created to select articles specifically relevant to GEO or PDB. Specifically, a rough selection of articles relevant for the specific databases that we studied was performed using naïve filtering on the corresponding evidence sentences: for GEO, articles were considered specifically relevant for GEO curation when the evidence sentence contained an occurrence of GEO, GSE or Gene Expression Omnibus. For PDB, articles were considered specifically relevant for PDB curation when the evidence sentence contained an occurrence of PDB, Protein Data Bank, Protein DataBank or RCSB.
The pool of these articles provided an evidence statement to support the fact that the article is of interest for data curation and was divided into three sets based on intersection with curated articles. Type ‘I’ sets represent the articles with evidence statements curated only by MEDLINE (SI), type ‘II’ sets represent the articles with evidence statements curated only by the biological database (GEO or PDB) and type ‘III’ sets represent the articles with evidence statements that were not curated, but only suggested, by the automatic tool.
Text-mining tool contribution to difference analysis
Automatic retrieval of evidence statements to support link curation
To perform a finer-grained analysis of curation differences, the text-mining tool allowed us to automatically retrieve specific evidence statements that could support the curated links found in the databases. The retrieved statements were sorted into three categories of interest as shown in Figures 3 and 4. To evaluate the quality of evidence statements, sets of 100 randomly selected statements were created for each category I, II and III for both GEO and PDB. The evidence statements were manually reviewed by two independent annotators (the authors worked in pairs) as outlined previously in the article.
The quality of evidence statements is consistently high
Table 3 shows the distribution of annotated categories for each set of articles with evidence statements. On an average, the inter-annotator agreement (measured as percentage of agreement) was high: 89% for GEO and 86% for PDB. The disagreements occurred mainly for statements that were annotated as ‘ambiguous’ by at least one annotator. As a result, the agreement was higher in sets that contained less of these ‘ambiguous’ statements.
Distribution of annotated categories for each set of articles with evidence statements
| Statement category . | GEO . | PDB . | ||||
|---|---|---|---|---|---|---|
| . | I . | II . | III . | I . | II . | III . |
| Deposition | 82 | 86 | 72 | 89 | 84 | 41 |
| Reuse | 4 | 4 | 13 | 2 | 3 | 24 |
| Ambiguous | 13 | 9 | 10 | 5 | 3 | 14 |
| Comment | 1 | 1 | 5 | 4 | 10 | 21 |
| Statement category . | GEO . | PDB . | ||||
|---|---|---|---|---|---|---|
| . | I . | II . | III . | I . | II . | III . |
| Deposition | 82 | 86 | 72 | 89 | 84 | 41 |
| Reuse | 4 | 4 | 13 | 2 | 3 | 24 |
| Ambiguous | 13 | 9 | 10 | 5 | 3 | 14 |
| Comment | 1 | 1 | 5 | 4 | 10 | 21 |
Distribution of annotated categories for each set of articles with evidence statements
| Statement category . | GEO . | PDB . | ||||
|---|---|---|---|---|---|---|
| . | I . | II . | III . | I . | II . | III . |
| Deposition | 82 | 86 | 72 | 89 | 84 | 41 |
| Reuse | 4 | 4 | 13 | 2 | 3 | 24 |
| Ambiguous | 13 | 9 | 10 | 5 | 3 | 14 |
| Comment | 1 | 1 | 5 | 4 | 10 | 21 |
| Statement category . | GEO . | PDB . | ||||
|---|---|---|---|---|---|---|
| . | I . | II . | III . | I . | II . | III . |
| Deposition | 82 | 86 | 72 | 89 | 84 | 41 |
| Reuse | 4 | 4 | 13 | 2 | 3 | 24 |
| Ambiguous | 13 | 9 | 10 | 5 | 3 | 14 |
| Comment | 1 | 1 | 5 | 4 | 10 | 21 |
For categories I and II, the number of statements classified as ‘deposition’ is very high, as could be expected from a curated source. However, a few statements are classified as ‘reuse’, pointing out some possible curation errors or cases where the adequate evidence statement was not found by the text-mining tool.
For category III, the number of statements classified as ‘deposition’ is lower than for categories I and II, resulting in lower inter-annotator agreement. The lower proportion of ‘deposition’ statements in this category could be expected, as contrary to category I and II, the fact that the articles are of interest for data curation is a result of automatic analysis alone and not supported by biological or literature database curators. In other words, these numbers reflect the free-range performance of the automatic tool. In this respect, two important points can be made: first, in terms of curation support, the tool can be assessed as highly useful because it provides evidence statements that can directly lead to a curation decision in 85% of cases for GEO (72 ‘deposition’ statements leading to positive curation decision and 13 ‘reuse’ statements leading to a negative curation decision) and 65% of cases for PDB (41 ‘deposition’ statements leading to positive curation decision and 24 ‘reuse’ statements leading to a negative curation decision). Second, in terms of tool performance across databases, these results indicate a better performance on GEO over PDB.
Discussion
Comparison to other work
Few studies have addressed the automatic extraction of links between biological databases and the literature using full text. To our knowledge, there is no existing work that can be directly compared with ours. The text-mining method applied here [first introduced in (5)] differs from other work (26,29) in at least the following three ways: (i) it makes the distinction between newly deposited data versus data reuse or data comment, (ii) it provides evidence statements to support fast curator decisions and (iii) it is database independent, so that it may be adopted to many contexts beyond that of GEO and PDB.
To our knowledge, this is the first study to compare the curation of links between biological databases and literature databases. It shows that in spite of a similar curation objective (curate links between biological database entries and articles in the literature describing the production of the data sets) the results, in terms of links, curated are different. This may be explained by the curation protocols used by MEDLINE and biological databases, both of which rely primarily on author-reported information. As authors contact the biological versus literature databases at different stages in the data deposition process, it can be hypothesized that the information available to them at these stages is different.
State of MEDLINE, GEO and PDB curation
This study shows that there are significant discrepancies between the curation of links between data and the literature found in MEDLINE and GEO and MEDLINE and PDB, respectively. For a better understanding of these differences, we have used a text-mining tool to retrieve evidence statements supporting the curation decisions. Several samples of evidence statements have been manually analysed to assess curation quality (sets of categories I and II in Table 3) and the quality of evidence statements supplied by the text-mining tool (sets of category III in Table 3). By extrapolating these results, we can estimate the error rate and the silence of link curation in each database as shown in Table 4. Error rate can be computed as E = 1−P, where P is precision. Silence can be computed as S = 1−R, where R is recall. Precision can be computed as the number of correctly curated links over the total number of curated links. Recall can be computed as the number of correctly curated links over the total number of links that should be curated.
Estimated error rate and silence of link curation in GEO, PDB and MEDLINE
| Steps to computation of silence and error rate . | GEO . | PDB . | MEDLINE (GEO) . | MEDLINE (PDB) . |
|---|---|---|---|---|
| Correctly curated links | 12 339 | 30 410 | 3135 | 19 937 |
| Curated links | 13 891 | 32 943 | 3208 | 20 286 |
| Links that should be curated | 18 865 | 35 953 | 18 865 | 35 953 |
| Precision | 88.8% | 92.3% | 97.7% | 98.3% |
| Recall | 65.4% | 84.6% | 16.6% | 55.5% |
| Error rate | 11.2% | 7.7% | 2.3% | 1.7% |
| Silence | 34.6% | 15.4% | 83.4% | 44.5% |
| Steps to computation of silence and error rate . | GEO . | PDB . | MEDLINE (GEO) . | MEDLINE (PDB) . |
|---|---|---|---|---|
| Correctly curated links | 12 339 | 30 410 | 3135 | 19 937 |
| Curated links | 13 891 | 32 943 | 3208 | 20 286 |
| Links that should be curated | 18 865 | 35 953 | 18 865 | 35 953 |
| Precision | 88.8% | 92.3% | 97.7% | 98.3% |
| Recall | 65.4% | 84.6% | 16.6% | 55.5% |
| Error rate | 11.2% | 7.7% | 2.3% | 1.7% |
| Silence | 34.6% | 15.4% | 83.4% | 44.5% |
Estimated error rate and silence of link curation in GEO, PDB and MEDLINE
| Steps to computation of silence and error rate . | GEO . | PDB . | MEDLINE (GEO) . | MEDLINE (PDB) . |
|---|---|---|---|---|
| Correctly curated links | 12 339 | 30 410 | 3135 | 19 937 |
| Curated links | 13 891 | 32 943 | 3208 | 20 286 |
| Links that should be curated | 18 865 | 35 953 | 18 865 | 35 953 |
| Precision | 88.8% | 92.3% | 97.7% | 98.3% |
| Recall | 65.4% | 84.6% | 16.6% | 55.5% |
| Error rate | 11.2% | 7.7% | 2.3% | 1.7% |
| Silence | 34.6% | 15.4% | 83.4% | 44.5% |
| Steps to computation of silence and error rate . | GEO . | PDB . | MEDLINE (GEO) . | MEDLINE (PDB) . |
|---|---|---|---|---|
| Correctly curated links | 12 339 | 30 410 | 3135 | 19 937 |
| Curated links | 13 891 | 32 943 | 3208 | 20 286 |
| Links that should be curated | 18 865 | 35 953 | 18 865 | 35 953 |
| Precision | 88.8% | 92.3% | 97.7% | 98.3% |
| Recall | 65.4% | 84.6% | 16.6% | 55.5% |
| Error rate | 11.2% | 7.7% | 2.3% | 1.7% |
| Silence | 34.6% | 15.4% | 83.4% | 44.5% |
In practice, we estimate the number of correctly curated links for a database, as the number of links curated in common with another database added to the number of links curated solely by that database weighted by the proportion of deposition for the corresponding category (I or II) in Table 3. For GEO, that number would be computed as 2805 + 86% × (13 891−2805), resulting in 12 339. We estimate the total number of links that should be curated as the number of uncurated links weighted by the proportion of deposition for category III in Table 3. The total number of uncurated links can be estimated proportionally with respect to curated versus uncurated citations in PMC. For GEO, curated versus uncurated citations in PMC amount to 5269 [PMC_GEOfilter ∩ (G2M U M2G), NPMIDs = 5269] and 2366, respectively. There are a total of 14 256 curated citations in MEDLINE, so the total number of uncurated citations can be estimated to be 6402 (14 256 × 2366 ÷ 5269). According to category III in Table 3, 72% of these uncurated articles (4609) actually contain deposition sentences. Finally, we add 4609 to 14 256 bringing the total number of links that should be curated to 18 865. Therefore, for GEO, precision can be computed as 12 339 ÷ 13 891, resulting in 88.8% (error rate is 1.2%) and recall can be computed as 12 339 ÷ 18 865, resulting in 65.4% (silence is 34.6%). Table 4 provides similar estimations for each link source in this study.
Recommendations for improved curation practices
The results of this study indicate several steps that may be undertaken to improve the curation of links between biological databases and the literature, involving different actors in the database maintenance and development process.
(5) Recommendation to journal editors: encourage authors to use non-ambiguous statements when reporting data deposition. For example, statements including the verbs ‘deposit’ or ‘submit’ are likely to be non-ambiguous, compared with statements using the adjective phrase ‘available’.
(6) Recommendation to database curators:
MEDLINE: Consider that deposition statements may occur in many sections of an article; in a footnote at the beginning, in a separate section at the end (currently considered by MEDLINE indexers), but also in the methods or results sections (not currently considered by MEDLINE indexers).
MEDLINE and biological database: The text-mining tool can provide assistance to curators by identifying evidence statements from full-text articles of interest (e.g. articles selected for biological database curation). The tool can also provide further automatic recommendations of articles to consider for (SI) indexing. Furthermore, when curation policies are similar (e.g. GEO and MEDLINE) the sharing of links could increase curation coverage and consistency between sources.
(7) Recommendation for authors: Curators rely heavily on author-reports for link curation. Therefore, authors are advised to follow-up on metadata reported to databases to ensure proper curation of their data sets and wider dissemination of their work.
These results and analysis have been shared with MEDLINE curators at NLM, who are currently planning to act on some of our recommendations, including the use of the text-mining tool to improve the coverage of links curated in the (SI) field for GEO and PDB.
Limitations of this study
The distribution of annotated categories for articles with evidence statements in set III (see Table 3) shows a better performance of the tool for GEO over PDB. This is consistent with our previous observations (5) and reflects the fact that an emphasis was given to microarray data (vs. protein structure or other types of data) in training the tool. Although the tool is not restricted to a specific biological data type, this indicates that for improved performance on a variety of data types, the training set should be augmented to include additional examples of data deposition statements beyond microarrays.
Furthermore, the automatic extraction of evidence statements is limited to the availability of full-text articles, as evidence statements mostly occur in full-text versus abstract. In this respect, there are variations between databases: for GEO, out of the 14 256 articles curated for links in either GEO or MEDLINE 6692 (47%) are available from PMC, whereas for PDB, out of the 35 211 articles curated for links in either PDB or MEDLINE 7904 (22%) are available from PMC.
Future work
In future work, we plan to follow-up with MEDLINE curators to systematically supply them with recommendations and evidence statements integrated in the NLM data creation and maintenance system used to update and maintain the MEDLINE database. This could provide us with curator feedback on the recommendations and evidence statements, by confronting the recommendations made with the curator decision as evidenced in the final citations. These judgments could be then used to improve on the quality of evidence statements supplied.
Conclusions
In this article, we have presented a comparative analysis of links between biological databases and the literature as curated by different sources. In spite of similar curation guidelines followed by GEO, MEDLINE and PDB curators, we find that the overlap between link sources is <50%. In addition, our analysis shows that links curated by only one source are relevant. To improve the consistency and coverage of all link sources, we propose the use of a text-mining tool to be able to process full-text articles to provide curators with recommendations of links to be curated, supported by evidence statements automatically extracted from full-text.
Funding
Intramural Research Program of the National Institutes of Health and the National Library of Medicine [LM000002-01].
Availability
The data sets described in this study will be made available freely on the NCBI website and as supplementary data.
Conflict of interest. None declared.
Acknowledgements
The authors thank the GEO and MEDLINE curators for helpful discussions on curation practices and data processing.