-
PDF
- Split View
-
Views
-
Cite
Cite
Ritu Khare, John D. Burger, John S. Aberdeen, David W. Tresner-Kirsch, Theodore J. Corrales, Lynette Hirchman, Zhiyong Lu, Scaling drug indication curation through crowdsourcing, Database, Volume 2015, 2015, bav016, https://doi.org/10.1093/database/bav016
Close - Share Icon Share
Abstract
Motivated by the high cost of human curation of biological databases, there is an increasing interest in using computational approaches to assist human curators and accelerate the manual curation process. Towards the goal of cataloging drug indications from FDA drug labels, we recently developed LabeledIn, a human-curated drug indication resource for 250 clinical drugs. Its development required over 40 h of human effort across 20 weeks, despite using well-defined annotation guidelines. In this study, we aim to investigate the feasibility of scaling drug indication annotation through a crowdsourcing technique where an unknown network of workers can be recruited through the technical environment of Amazon Mechanical Turk (MTurk). To translate the expert-curation task of cataloging indications into human intelligence tasks (HITs) suitable for the average workers on MTurk, we first simplify the complex task such that each HIT only involves a worker making a binary judgment of whether a highlighted disease, in context of a given drug label, is an indication. In addition, this study is novel in the crowdsourcing interface design where the annotation guidelines are encoded into user options. For evaluation, we assess the ability of our proposed method to achieve high-quality annotations in a time-efficient and cost-effective manner. We posted over 3000 HITs drawn from 706 drug labels on MTurk. Within 8 h of posting, we collected 18 775 judgments from 74 workers, and achieved an aggregated accuracy of 96% on 450 control HITs (where gold-standard answers are known), at a cost of $1.75 per drug label. On the basis of these results, we conclude that our crowdsourcing approach not only results in significant cost and time saving, but also leads to accuracy comparable to that of domain experts.
Database URL: ftp://ftp.ncbi.nlm.nih.gov/pub/lu/LabeledIn/Crowdsourcing/ .
Introduction
A common task in biocuration is to manually extract knowledge from unstructured texts and transform them into structured datasets. Manual data curation is very central to the contemporary biomedical research, as it generates computable data that is accessible to both machines and end users. However, manual curation is inherently expensive due to the associated time and human labor costs. In response to the scalability issue of manual curation, recently there has been an increasing interest in using advanced computer technologies for assistance, including various text-mining techniques ( 1 , 2 ) and interactive computer systems ( 3–5 ). To our knowledge, existing efforts have been primarily focused on improving biocuration workflows ( 6–8 ) and common literature curation tasks such as document triage ( 9 , 10 ), gene tagging ( 11 ), and Gene Ontology (GO) annotation ( 12 , 13 ).
Unlike previous studies, the ultimate goal of this work is to curate medical information, more specifically therapeutic relationships between human drugs and diseases, from the free text descriptions into structured knowledge. Previous research suggests that such a structured and computable resource is critical for many real-world applications, ranging from online health information retrieval ( 14–16 ), to translational bioinformatics research ( 17–20 ), to clinical decision support systems ( 21–23 ). Given the lack of such a gold standard, there have been several attempts ( 24–26 ) towards creating a comprehensive repository of drug–disease relationships in the public domain. For such a purpose, the drug Structured Product Labeling (SPL) data (hereafter referred to as drug labels; see Figure 1 for an example) has been more commonly used than the biomedical literature. Drug labels contain rich textual descriptions of drug indications and clinical trial studies for marketed drugs. They are submitted to the FDA by the pharmaceutical manufactures and can be freely downloaded from the U.S. National Library of Medicine’s DailyMed http://dailymed.nlm.nih.gov/dailymed/index.cfm database.

An example of an FDA Drug Label in DailyMed; drug names are specified as normalized concepts under the ‘RxNorm Names’ box, and the drug indications are described as free text in the ‘INDICATIONS AND USAGE’ section.
Towards such a goal, we recently created LabeledIn ( 27 ) based on manual curation of drug labels. To accelerate the manual curation process, we adopted a semiautomated pipeline where all disease occurrences are first tagged by a text-mining tool. Next, human experts were asked to select true indications and reject non-indication disease mentions. The manual annotation process involved three highly experienced annotators with expertise in pharmacy and biomedical document indexing, with the assistance of detailed annotation guidelines http://ftp.ncbi.nlm.nih.gov/pub/lu/LabeledIn/Annotation_Guidelines.pdf . For cataloging indications of 250 popular human drugs, it effectively required over 40 h of human labor, spread across over 20 weeks. With our ultimate goal to scale LabeledIn with thousands of drugs from DailyMed and other resources, this study investigate the feasibility of scaling human curation through a technique of crowdsourcing, and subsequently evaluate its efficiency, cost-effectiveness and ability to achieve high-quality annotations.
Crowdsourcing is known as a participative online activity wherein a job of variable complexity and modularity is outsourced to an undefined, diverse, and large network of workers ( 28 ). Over the years, its definition has evolved to include many activities, such as analysis of search logs ( 14 ), editing wikis in biology ( 29 ), etc. The chief characteristic of a task to be formulated as a crowdsourcing job is that it cannot be solved by automated computational methods. Good and Su ( 30 ) focus on directed problems in bioinformatics and classify the crowdsourcing problems in biomedicine as (i) megatasks, tasks that are individually challenging, e.g. open innovation contests ( 31 ), and (ii) microtasks, tasks that are large in number but low in difficulty, e.g. word sense disambiguation ( 32 ) and named entity recognition ( 33 ). The workers participate in microtasks for a variety of reasons, such as altruism (science service), fun (scientific games), cash rewards (microtask markets), or out of necessity (the ReCAPTHA project for optical character recognition).
With regard to the task of text annotation, the microtask markets powered by the Amazon Mechanical Turk or MTurk ( https://www.mturk.com ) platform are among the most popular crowdsourcing mechanisms. In microtasking with MTurk, a crowdsourcing job is decomposed into modular units known as a human intelligence task (HIT): a task that can be quickly accomplished by humans (referred to as ‘turkers’) who are generally under age 30 and have a college or advanced degree ( 34 ). Successful uses of MTurk for producing linguistic resources or performing evaluations at a low cost have been reported in the natural language processing (NLP) domain. Two biomedical microtasking studies include the medical named entity recognition (NER) and linking study ( 35 ) and the gene–mutation relationship study ( 36 , 37 ). But unlike previous NLP-oriented tasks, we aim to tackle a problem in biomedical data curation rather than linguistic or corpus annotation with very different objectives and practices (e.g., our crowdsourced results neither include any linguistic information nor are meant for training or evaluating NLP algorithms). Thus our study has more resemblance to ( 23 , 24 ) than ( 22 ). But compared to ( 22 ), we were able to achieve higher accuracies on this task with several innovations in design and quality control.
Motivated by the previous success, we also rely on the microtask market through MTurk but explore its use on a new problem (curating indications from drug labels) in biomedicine as opposed to those traditional tasks (e.g. entity tagging). More specifically, we reformulate the complex task of annotating drug–disease treatment relationships (by domain experts as in LabeledIn) into HITs suitable for the average turkers in the microtasking environment. In creating LabeledIn, an expert curator was shown all pre-computed disease mentions in a drug label and asked to select the correct indication(s) and reject invalid ones. In contrast, in this study the annotation task is significantly simplified: only one disease is shown in a HIT at a time to the turkers such that a binary YES/NO judgment will be sufficient. Next, a novel feature of our HIT design is that to simulate the guided nature of expert annotation studies, we encode the lengthy annotation guidelines as multiple-choice answers in a HIT. That is, in order to assist turkers to reject non-indication disease mentions in a drug label, we expanded one NO option into five, each representing a specific reason for rejection (e.g. the highlighted disease is a side effect of the drug). Then in order to optimize the quality of crowdsourced results, we implemented a number of quality control measures such as using a high cut-off qualifier test and representative control items. Finally, the scale of this study (3454 HITs drawn from 793 drug labels) is comparable to the other MTurk studies in biomedicine. Our experimental results, including turnaround time and aggregated HIT accuracy, strongly indicate the practical utility of crowdsourced judgments in increasing the drug coverage of LabeledIn.
Methods
Figure 2 shows our overall framework of cataloging drug–disease relationships from FDA drug labels using MTurk in five modules:
Select the drug labels from DailyMed.
Use NER tools to tag all diseases and drugs in a given drug label.
Design and generate atomic tasks, or HITs.
Configure the MTurk platform and submit the HITs for crowdsourcing.
Collect, aggregate, and evaluate the turkers’ judgments (based on the control items).

Crowdsourced Microtasking Pipeline for Cataloging Drug Indications from FDA Drug Labels. Part II shows the drug and disease mentions identified using named-entity recognition (NER) tools.
The first two modules are adopted from the original LabeledIn study pipeline, wherein identical drug labels are grouped to minimize overall workload, and NER tools are applied to aid the annotation process. For the remaining modules, we translate the task of domain expert annotation into microtasks for use within the environment and technical limitations associated with MTurk. We designed this study and prepared the datasets for crowdsourcing within a span of 3 months.
Part I. Drug label selection
DailyMed contains drug SPL data for many kinds of medications such as human prescription drugs, over-the-counter drugs, animal drugs, vaccines, homeopathic drugs, etc. In this study, we focus on human prescription and over-the-counter drugs. We downloaded the August 2012 release of DailyMed, which contains 28 178 applicable drug labels, i.e. drug labels that have non-empty indication and drug concept sections. These drug labels are associated with 2025 different ingredients as computed from the linked RxNorm names (RxNorm is a standardized nomenclature for clinical drugs produced by the National library of Medicine: http://www.nlm.nih.gov/research/umls/rxnorm/ ) and identifiers (see for example the green box in Figure 1 ).
Our drug label selection process takes into account the popularity of drugs in terms of their online access in PubMed Health ( http://www.ncbi.nlm.nih.gov/pubmedhealth/ ). Earlier in LabeledIn ( 27 ), we included 8151 drug labels corresponding to 250 highly accessed drugs (e.g., Alprazolam, Citalopram) accounting for 72.81% of the total drug access on PubMed Health. In this study, through crowdsourcing, we aim to increase the coverage of LabeledIn to 95% of total drug access. Accordingly, we identified 276 drugs that account for an additional 22.19% of total drug access. These 276 drugs correspond to 6488 drug labels (many are redundant due to submissions by different pharmaceutical manufacturers).
To minimize annotation workload and avoid information loss downstream, we adopted a drug label grouping mechanism ( 27 ), where we group the drug labels having almost identical textual descriptions together, and choose a representative from each group for human annotation. In particular, we considered two drug labels to be identical if their Dice coefficient measure was above a threshold of 0.87 (empirically determined). In this manner, we reduced the 6488 drug labels to 706 unique drug labels. Finally, for these 706 drug labels, we extract their indication sections and use them as input for the next part of the framework. We also maintain the information about the linked drug concepts (in RxNorm identifiers) for these drug labels.
Part II. Named entity recognition
We utilize the high-recall disease NER method from our previous work ( 27 ) to highlight all disease names in a given drug label. In addition, we also highlight the relevant drug names in the drug labels. Figure 2 (Part II) shows the results of applying our NER modules on a drug label.
Disease NER
We used MetaMap ( 38 ) to tag all disease names mentioned in the indication section of the drug labels. In the LabeledIn study ( 27 ), we configured MetaMap to deliver a high recall on disease mentions and achieved 94% recall on 500 drug labels. We limited the semantic types to the disorder semantic group in the UMLS, and the source vocabularies to SNOMED CT and MeSH, previously found to be useful in annotating clinical documents and biomedical research articles, respectively ( 39–42 ). We also take advantage of other MetaMap features such as word sense disambiguation, term processing and Metathesaurus candidates. Finally, we obtain the disease mentions from the drug labels, including their corresponding offsets and UMLS identifiers (i.e. CUIs). Overall, 3004 <drug-label, disease-CUI> pairs were returned using this method. In addition, we further refine the NER results by including the disease abbreviations identified using an abbreviation resolution tool ( 43 ), e.g. ‘MDD’ for ‘major depressive disorder.’
Drug NER
We also tag the drug names mentioned in the drug label using a lexical method based on RxNorm: Since each drug label is already linked to a list of precise RxNorm drug concepts (see an example in Figure 1 ), we simply derive a list of active ingredients and brand names in RxNorm associated with each drug label. We then use a dictionary look-up method to identify the occurrences of these names in the drug label. Note that for each drug label, we only identify and highlight the drug names associated with the linked drug concepts.
Part III. Human intelligence task (HIT) design and generation
The main challenge, thereafter, was to translate the complex task of expert annotation into manageable atomic units, aka, HITs that can be solved by an unknown network of turkers. We randomly selected 95 drug labels out of 500 drug labels previously annotated in LabeledIn, and used the MTurk Developer Sandbox tool to conduct a pilot study to iteratively brainstorm and study various design alternatives among the investigators of this study.
The first key outcome of our pilot study was to display one disease concept at a time (instead of showing all diseases at once) in a single HIT. This is in contrast to the original expert-annotation pipeline where all diseases are shown together (e.g., three different disease concepts in the drug label in Figure 1 ) and the domain experts were required to make multiple judgments at once within a single drug label; this often led to disagreements or errors especially in case of dense labels ( 26 ). This design decision is also motivated by the previous crowdsourcing study ( 36 , 37 ) and ensures that one HIT corresponds to making only one decision, i.e. to judge whether the drug in question is used in managing the highlighted disease. Figure 3 demonstrates that three HITs are generated for the drug label in Figure 2 . Note that the two mentions, ‘major depressive disorder’ and ‘depression,’ correspond to two different disease concepts, and hence translate into separate HITs.
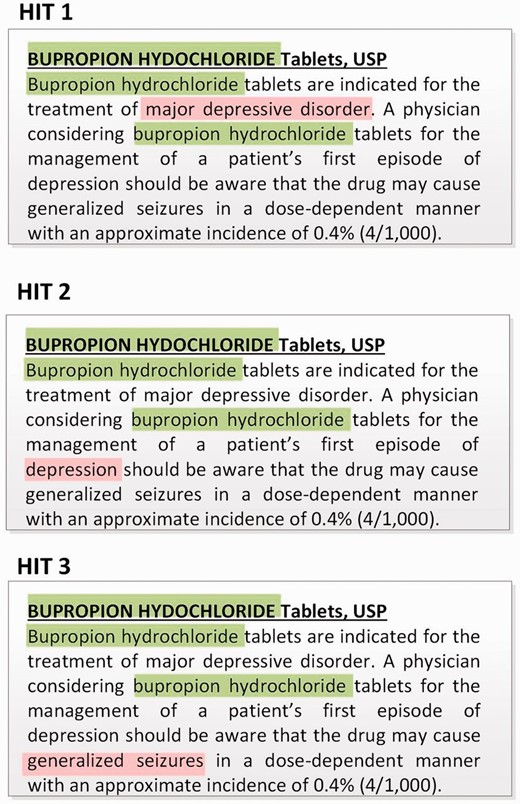
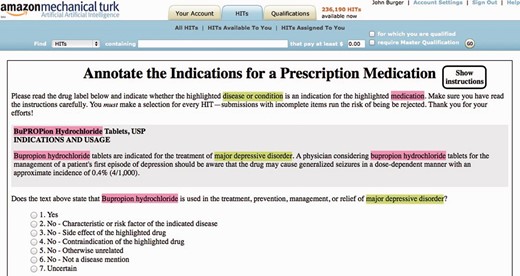
The second key outcome of the pilot study is motivated by the fact that in the previous LabeledIn study ( 27 ), the domain experts relied heavily on the standalone annotation guidelines (i.e. not part of the annotation interface) to make their judgments based on multiple inclusion (i.e. what to annotate) and exclusion (i.e. what not to annotate) criteria. In the crowdsourcing interface, we initially incorporated all these guidelines into the instructions. However, in the pilot study we found it very inconvenient to frequently switch back and forth between the actual HIT and the instructions. We realized that this would not only slow down the process but also discourage turkers from pursuing our crowdsourcing job correctly. Hence, we incorporated the annotation guidelines into the HIT itself, in particular, by embedding major scenarios into user options while still keeping the number of options manageable for human processing ( 44 ). Specifically, we designed the HIT question to facilitate turkers in thinking about reasons for rejecting a certain disease as an indication, making this problem a six-way classification task as shown in Figure 4 (see bottom; an additional option is also reserved for users with uncertain answers but is rarely used in our results). Table 1 shows an example of each of the five categories of non-indication diseases mentioned in drug labels corresponding to the NO user option. Note that unlike the usual multi-way classification task for capturing different classes of answers, our design of six categories specifically intends to facilitate users in ultimately making correct binary YES or NO judgments.
| Category . | Example . |
|---|---|
| Characteristic or risk factor | Doxazosin mesylate is indicated for the treatment of both the urinary outflow obstruction and obstructive and irritative symptoms associated with BPH: obstructive symptoms (hesitation, intermittency, dribbling , weak urinary stream, incomplete emptying of the bladder) |
| Side effect | A physician considering bupropion hydrochloride tablets for the management of a patient’s first episode of depression should be aware that the drug may cause generalized seizures in a dose-dependent manner with an approximate incidence of 0.4% |
| Contraindication | Carbamazepine is not a simple analgesic and should not be used for the relief of trivial aches |
| Unrelated | Ranitidine is indicated in the treatment of GERD. Concomitant antacids should be given as needed for pain relief to patients with GERD |
| Not a disease | Promethazine hydrochloride tablets are useful for the prevention and control of nausea and vomiting associated with certain types of anesthesia and surgery |
| Category . | Example . |
|---|---|
| Characteristic or risk factor | Doxazosin mesylate is indicated for the treatment of both the urinary outflow obstruction and obstructive and irritative symptoms associated with BPH: obstructive symptoms (hesitation, intermittency, dribbling , weak urinary stream, incomplete emptying of the bladder) |
| Side effect | A physician considering bupropion hydrochloride tablets for the management of a patient’s first episode of depression should be aware that the drug may cause generalized seizures in a dose-dependent manner with an approximate incidence of 0.4% |
| Contraindication | Carbamazepine is not a simple analgesic and should not be used for the relief of trivial aches |
| Unrelated | Ranitidine is indicated in the treatment of GERD. Concomitant antacids should be given as needed for pain relief to patients with GERD |
| Not a disease | Promethazine hydrochloride tablets are useful for the prevention and control of nausea and vomiting associated with certain types of anesthesia and surgery |
| Category . | Example . |
|---|---|
| Characteristic or risk factor | Doxazosin mesylate is indicated for the treatment of both the urinary outflow obstruction and obstructive and irritative symptoms associated with BPH: obstructive symptoms (hesitation, intermittency, dribbling , weak urinary stream, incomplete emptying of the bladder) |
| Side effect | A physician considering bupropion hydrochloride tablets for the management of a patient’s first episode of depression should be aware that the drug may cause generalized seizures in a dose-dependent manner with an approximate incidence of 0.4% |
| Contraindication | Carbamazepine is not a simple analgesic and should not be used for the relief of trivial aches |
| Unrelated | Ranitidine is indicated in the treatment of GERD. Concomitant antacids should be given as needed for pain relief to patients with GERD |
| Not a disease | Promethazine hydrochloride tablets are useful for the prevention and control of nausea and vomiting associated with certain types of anesthesia and surgery |
| Category . | Example . |
|---|---|
| Characteristic or risk factor | Doxazosin mesylate is indicated for the treatment of both the urinary outflow obstruction and obstructive and irritative symptoms associated with BPH: obstructive symptoms (hesitation, intermittency, dribbling , weak urinary stream, incomplete emptying of the bladder) |
| Side effect | A physician considering bupropion hydrochloride tablets for the management of a patient’s first episode of depression should be aware that the drug may cause generalized seizures in a dose-dependent manner with an approximate incidence of 0.4% |
| Contraindication | Carbamazepine is not a simple analgesic and should not be used for the relief of trivial aches |
| Unrelated | Ranitidine is indicated in the treatment of GERD. Concomitant antacids should be given as needed for pain relief to patients with GERD |
| Not a disease | Promethazine hydrochloride tablets are useful for the prevention and control of nausea and vomiting associated with certain types of anesthesia and surgery |
Finally, based on the pilot study, we decided to also highlight the drug mentions in order to provide better readability of the HITs, unlike the previous LabeledIn study where we only highlighted the disease mentions. For the drug labels with no occurrences of drug names in their indication field, we simply added a title with its generic name highlighted above the indication field.
Part IV. The crowdsourcing job
Our crowdsourcing experimental dataset includes 706 FDA drug labels corresponding to 276 ingredients as described in the ‘Part I. Drug Label Selection’ section. The total number of corresponding HITs was 3004, i.e. equal to the number of <drug-label, disease-CUI> pairs identified in the ‘Part II. Disease NER’ subsection. A HIT screenshot of the MTurk interface is shown in Figure 4 .
Before posting the HITs on MTurk, we implemented several quality-control measures as recommended by earlier studies ( 35–37 ). Since the SPL data represents the drug package inserts for U.S. consumers, we restricted the job to turkers from the US with English language ability, although we are aware that this cannot be guaranteed ( 45 ). We designed a 10 HIT-qualifier test, drawn from the gold standard in LabeledIn, to ensure that only the turkers with clear understanding of the task and the associated instructions are permitted to work on our HITs. The 10 test cases are distributed into 4 YES and 6 NO HITs, drawn from different NO categories of the annotation guidelines. We only allowed turkers obtaining 80% or higher score on this test to proceed further. This high cut-off also ensures that the English language requirement is met given the variety of language descriptions in the test cases.
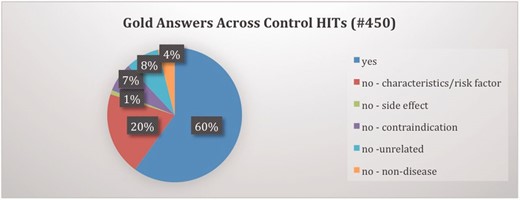
To measure the performance of turkers, we utilized the control item feature of MTurk wherein some HITs with known or gold answers are inserted after every few new HITs. We were able to utilize this critical feature because of the availability of our previous expert-annotated LabeledIn dataset. We selected a subset of LabeledIn: 450 disease concepts in 87 drug labels with 60/40 distribution of YES/NO judgments. We had to re-annotate the NO judgments in this LabeledIn subset to further classify them into one of the five categories as shown in Table 1 . Figure 5 shows the distribution of gold answers across the control items. We configured this study such that the 450 control HITs accounted for approximately 20% of total HITs.
Part V. Judgment collection and consensus building
Finally, we employed redundancy of judgments and accepted five judgments per HIT. We paid six cents per HIT to each turker based on a small cost determination experiment. The full MTurk study, for collecting judgments for 3004 new HITs and 450 control HITs, cost $1239.15. For aggregating turkers’ judgments, we used two commonly used methods: majority voting and expectation maximization ( 46 , 47 ). The majority-voting scheme makes the assumption that every turker is equally accurate and takes the majority class among all judgments as the adjudicated judgment.
The expectation maximization algorithm (hereafter referred to as the EM algorithm) was proposed to address a mixture of expertise levels that is often observed in crowdsourced labels. It is an iterative method used to determine maximum likelihood parameters with latent or unknown variables. In our task, the gold standard labels (YES or NO) are the unknown values, while we estimate each annotator’s sensitivity and specificity in order to maximize the likelihood of seeing our data. For this study, we implemented the version described in Raykar et al.( 46 ). The EM algorithm’s first iteration is equivalent to majority voting, except that it retrieves soft labels (i.e. 3/5 yes votes is a soft label of 0.6 for the HIT). It then uses these labels to update each turker’s sensitivity and specificity, where each one is a weighted average of that turker’s responses. Soft labels are then recalculated using the Bayes theorem given the turker’s labels and sensitivity/specificity estimates. Essentially, the algorithm measures each turker against its current best guesses for the true labels, and assigns accuracy values based on how often the turker matches its estimate. In both methods, uncertain judgments (option #7 in Figure 4 ) were discarded.
Evaluation
We computed the accuracy of the crowdsourced judgments on the 450 control HITs (gold answers are available) at different levels: First, we computed the judgment-wise accuracy: average accuracy of each judgment against the gold standard. Next, we computed the turker-wise accuracy: the average accuracy of individual turkers. We not only calculate this for all turkers, but also for prolific turkers (e.g. turkers who worked on more than 50 control HITs, i.e. 50+ Turker Accuracy). Finally, we reported HIT-wise accuracies for both aggregation methods. Although our ultimate goal is to assess accuracy of binary classification (YES or NO), we also compute results of six-way classification where judgments of different NO categories are used separately.
Results
The drug indication crowdsourcing job was finished in 7 h and 58 min of job posting. A total of 113 turkers took the qualifier test, out of which 92 passed the test. A total of 74 turkers worked on at least one HIT, and 64 turkers worked on at least one control item. Overall, 18 775 judgments, including 3755 control item judgments, were collected through this job. The 3004 new HITs each received five judgments per our request. Among the 450 control HITs, each HIT received 5 or 10 judgments each, as per MTurk’s internal mechanism of feeding control HITs to turkers. When any turker worked on the same item more than once, we preferred their most recent judgment, leading to 3470 control item judgments. All 3470 judgments were used for judgment-wise and turker-wise accuracy calculations, but only 3417 were used for HIT-wise accuracy calculations because 53 uncertain judgments (1.53%) were not used in consensus building methods.
Table 2 shows the turker performance on the control items computed using various methods, and includes the number of applicable HITs, total judgments, total turkers, YES/NO accuracy, and six-way classification accuracy. The judgment-wise accuracy of the turkers was 90.95%. In general, the turker-wise accuracy increased along with the number of HITs submitted by a turker, and the most prolific turker achieved 93.25% accuracy. The EM algorithm, after 20 iterations achieved an accuracy of 95.78%. We stopped updates after 20 iterations as there were no more changes in the estimated results thereafter. The majority voting across turkers achieved 96% accuracy.
| Method . | Number of HITs . | Number of Judgments . | Number of Turkers . | Yes/no accuracy (%) . | Six-way accuracy (%) . |
|---|---|---|---|---|---|
| Judgment wise | 450 | 3470 | 64 | 90.95 | 83.22 |
| Turker wise | 450 | 3470 | 64 | 88.39 | 81.14 |
| 50+ turker wise | 450 | 2953 | 26 | 91.54 | 83.98 |
| 100+ turker wise | 450 | 2252 | 15 | 90.21 | 82.83 |
| Most prolific turker | 341 | 341 | 1 | 93.25 | 85.63 |
| EM | 450 | 3417 | 64 | 95.78 | 88.44 |
| Majority voting | 450 | 3417 | 64 | 96.00 | 88.66 |
| Method . | Number of HITs . | Number of Judgments . | Number of Turkers . | Yes/no accuracy (%) . | Six-way accuracy (%) . |
|---|---|---|---|---|---|
| Judgment wise | 450 | 3470 | 64 | 90.95 | 83.22 |
| Turker wise | 450 | 3470 | 64 | 88.39 | 81.14 |
| 50+ turker wise | 450 | 2953 | 26 | 91.54 | 83.98 |
| 100+ turker wise | 450 | 2252 | 15 | 90.21 | 82.83 |
| Most prolific turker | 341 | 341 | 1 | 93.25 | 85.63 |
| EM | 450 | 3417 | 64 | 95.78 | 88.44 |
| Majority voting | 450 | 3417 | 64 | 96.00 | 88.66 |
| Method . | Number of HITs . | Number of Judgments . | Number of Turkers . | Yes/no accuracy (%) . | Six-way accuracy (%) . |
|---|---|---|---|---|---|
| Judgment wise | 450 | 3470 | 64 | 90.95 | 83.22 |
| Turker wise | 450 | 3470 | 64 | 88.39 | 81.14 |
| 50+ turker wise | 450 | 2953 | 26 | 91.54 | 83.98 |
| 100+ turker wise | 450 | 2252 | 15 | 90.21 | 82.83 |
| Most prolific turker | 341 | 341 | 1 | 93.25 | 85.63 |
| EM | 450 | 3417 | 64 | 95.78 | 88.44 |
| Majority voting | 450 | 3417 | 64 | 96.00 | 88.66 |
| Method . | Number of HITs . | Number of Judgments . | Number of Turkers . | Yes/no accuracy (%) . | Six-way accuracy (%) . |
|---|---|---|---|---|---|
| Judgment wise | 450 | 3470 | 64 | 90.95 | 83.22 |
| Turker wise | 450 | 3470 | 64 | 88.39 | 81.14 |
| 50+ turker wise | 450 | 2953 | 26 | 91.54 | 83.98 |
| 100+ turker wise | 450 | 2252 | 15 | 90.21 | 82.83 |
| Most prolific turker | 341 | 341 | 1 | 93.25 | 85.63 |
| EM | 450 | 3417 | 64 | 95.78 | 88.44 |
| Majority voting | 450 | 3417 | 64 | 96.00 | 88.66 |
We performed further analysis of the 18 HITs where our best consensus building method (majority voting) differed from the gold standard answers. There were eight cases that could be considered ambiguous in that these scenarios were not described or explained in our instructions/examples, e.g. in the statement ‘Attention Deficit Disorders: Other terms being used to describe the behavioral syndrome include Hyperkinetic Child Syndrome, Minimal Brain Damage, …’ the turkers judged the ‘behavioral syndrome’ concept as YES as it was the only highlighted disease concept in the HIT. On the other hand, in our gold standard, this disease concept was specified as NO since a more specific concept (‘Attention Deficit Disorders’) was included as an indication. There were seven cases where the aggregated turker results were not correct, e.g. selecting ‘hyperthyroidism’ as an indication from ‘Thyroid hormone drugs are used as diagnostic agents in suppression tests to differentiate suspected mild hyperthyroidism or thyroid gland autonomy.’ Also, there were three cases where the gold standard was incorrect, raising our final accuracy to 96.67% effectively.
Discussion and conclusions
With the ultimate goal of scaling, the curation of a unique and computable resource on drug indications, this study uses the crowdsourcing microtask market to investigate whether accurate annotations could be achieved in a manner more efficient than contemporary expert-annotation pipelines. More specifically, we posted 3004 HITs corresponding to 706 new drug labels on the MTurk platform, and also injected 450 controls HITs drawn from previously annotated 87 drug labels to assess the turkers’ performances ( 35 ). All judgments were collected within 8 h of job posting, in contrast to over 40 h of human expert effort that was required to annotate 500 drug labels in our previous study ( 27 ). In addition to time saving, the crowdsourced judgments were collected at a minimal cost of $1.75 per drug label compared to the cost of $10 to $50 when expert curation is needed. Finally, compared to a small group of experts in traditional biocuration projects, this crowdsourcing study recruited 74 turkers offering more diversity in annotation ( 34 ).
Note that the 96% of accuracy in our results is also consistent with that of ( 36 ) where the turkers were 90% accurate on gene-mutation relation judgment. Our relatively higher performance in this study suggests that the drug indication curation task is perhaps more suitable for the crowdsourcing approach. After all, drug labels are for use by the general public while scientific articles are mostly written for professionals (e.g. researchers) to comprehend.
Despite success, several technical limitations of MTurk ( 48 ) previously noted in producing linguistic resources or performing NLP evaluations are worth noticing. For instance, several past studies show that it is difficult to have turkers perform complex tasks with results comparable to experts or standard machine learning techniques. Motivated by these earlier observations and our own experience of creating LabeledIn (a complex expert-annotation task), we took specialized simplification steps in designing the microtask itself (only a binary decision is required per HIT) and in the design of the HIT user interface (inserting annotation guidelines into the HIT interface). Together with other quality control measures (e.g. qualifier test), we achieved higher performance than that of an automatic classification method ( 49 ), which was also developed to address the scalability in drug indication annotation.
With regards to the ethical issues noted in the past research, we would like to add that we value ethics over cost savings, and are fully aware of the finding that the majority of active turkers rely on MTurk as a primary or secondary source of income ( 48 ). In our study, the workload for each HIT was kept minimal using several measures, and we approved the payments for all turkers who worked on at least one control HIT. We are also aware of the legal and ethical consequences of MTurk ( 50 ), but these issues are beyond the scope of this study.
Finally, we acknowledge several remaining issues in this study: First, there is the recall limitation of our NER tools despite our best efforts; improving the disease NER task remains a future task ( 38 , 51 ). Next, we would like to distinguish specific versus generic disease mentions in a drug label. Also, we plan to use the EM algorithm to estimate and select which HITs would most benefit from additional expert review (e.g. the HITs that received uncertain or conflicted judgments) whereby we can further improve of the quality of the crowdsourced judgments. Lastly, we acknowledge that the design of this study largely depends on the earlier study to produce LabeledIn that was designed and conducted in a span of 6 months, and the design of this crowdsourcing study was brainstormed and finalized in a span of 3 months. However, the time spent in the design of this study is a one-time effort as we could re-use the methodology for remaining drug labels and re-adapt for other drug indication sources.
In summary, we have introduced a crowdsourcing framework that recruits unknown workers to judge whether a highlighted disease is an indicated use for a given drug. Through our unique design of the crowdsourcing task and multiple quality control measures, we successfully demonstrated that it is feasible to achieve high-quality results using the MTurk platform for the drug indication curation task in a cost-effective and time-efficient manner.
Funding
The work was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine (R.K., Z.L.). T.C. was a summer intern at the NIH and supported by the NIH Intramural Research Training Award. The work was also funded under MITRE’s Internal Research and Development Program (J.A., J.B., D.T.K., L.H.). Funding for open access charge: The Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest . None declared.
References



