-
PDF
- Split View
-
Views
-
Cite
Cite
Soo-Yong Shin, Sun Kim, W. John Wilbur, Dongseop Kwon, BioC viewer: a web-based tool for displaying and merging annotations in BioC, Database, Volume 2016, 2016, baw106, https://doi.org/10.1093/database/baw106
Close - Share Icon Share
Abstract
BioC is an XML-based format designed to provide interoperability for text mining tools and manual curation results. A challenge of BioC as a standard format is to align annotations from multiple systems. Ideally, this should not be a major problem if users follow guidelines given by BioC key files. Nevertheless, the misalignment between text and annotations happens quite often because different systems tend to use different software development environments, e.g. ASCII vs. Unicode. We first implemented the BioC Viewer to assist BioGRID curators as a part of the BioCreative V BioC track (Collaborative Biocurator Assistant Task). For the BioC track, the BioC Viewer helped curate protein-protein interaction and genetic interaction pairs appearing in full-text articles. Here, we describe the BioC Viewer itself as well as improvements made to the BioC Viewer since the BioCreative V Workshop to address the misalignment issue of BioC annotations. While uploading BioC files, a BioC merge process is offered when there are files from the same full-text article. If there is a mismatch between an annotated offset and text, the BioC Viewer adjusts the offset to correctly align with the text. The BioC Viewer has a user-friendly interface, where most operations can be performed within a few mouse clicks. The feedback from BioGRID curators has been positive for the web interface, particularly for its usability and learnability.
Database URL : http://viewer.bioqrator.org
Background
As text mining has gained popularity in the biomedical domain, many biomedical natural language processing tools have been developed and released to the public. While many of these tools are useful, the difficulty comes from the integration with a user’s existing framework. This is due to the fact that relatively few tools support a common format that can be easily used for exchanging data. General frameworks are available to solve this interoperability issue ( 1 ); however, a drawback of these approaches is their steep learning curve. Thus, the new data exchange format, BioC ( 1 ), was developed through the BioCreative interoperability initiative ( 2 ). BioC is an XML-based format for embedding text, annotations and relations. Not only is BioC simple and straightforward, BioC programming libraries ( 1 , 3 ) are also freely available in multiple languages such as C ++, Java, Perl, Python, Go and Ruby. Moreover, there are a number of text mining and curation tools ( 4–8 ) that support BioC.
Though efforts have been made to enhance BioC, some hurdles yet remain. One issue is the misalignment of offsets between annotations and text in BioC files. In a BioC file, passages and sentences have offsets, i.e. passages (or sentences) start from given offsets in a document. Hence, the offset of an annotation is calculated as the sum of the passage (or sentence) offset that the annotation belongs to and the relative position of the annotation from the passage (or sentence) offset. Even if users follow the guidelines outlined by BioC key files ( 1 ), different character encodings, e.g. ASCII vs. Unicode, may still cause a misalignment problem. Until now, it has been difficult to solve misalignment issues because there has been no tool that can verify and possibly correct misaligned annotation offsets. Furthermore, there are no convenient tools available for merging BioC files. When users want to combine different types of annotations from multiple BioC files, they have to build their own merging tools. PubAnnotation ( 9 ), a persistent and sharable corpus and annotation repository, provides a tool that can handle both misalignment and annotation merger issues, but as of today, the interface does not support the BioC format. Argo ( 10 ) and Egas ( 7 ) can be utilized for solving these issues. However, Argo is a generic text mining platform and its user interface is not tailored for BioC. The Egas interface cannot be used to merge multiple BioC files.
The BioCreative V BioC track ( 11 , 12 ) is a continuing effort promoting the BioC format. The goal of the BioC track is to create BioC-compatible modules that complement each other and integrate into a system that assists BioGRID ( 13 ) curators. For the BioC track, we participated in Task 8 to create a visual tool for displaying various annotations. This task was to develop a visualization tool for highlighting protein–protein interaction (PPI) and genetic interaction (GI) annotations as well as gene/protein/organism mentions from text mining systems. A curation capability was also required to allow the recording of protein pairs with PPI and GI relationships. As a result, we developed the BioC Viewer ( http://viewer.bioqrator.org ), a web interactive tool for visualizing and curating PPI and GI information ( 14 ). This interface is based on our implementation of the PubMed ® abstract annotation tool, BioQRator ( 8 , 15 ). Unlike the previous interface, the BioC Viewer supports a display function for full-text articles, i.e. PubMed Central ® (PMC) articles. The curation tool for BioGRID curators is a plug-in function assisting PPI and GI curations. Since one of our main goals was to make an easy-to-use tool for curators, we designed the interface for users to complete most operations within a few mouse clicks.
While interacting with other participating teams and the organizers of the BioCreative V BioC track, we realized the necessity of a BioC validation tool. Even with the same corpus, the outcomes from different teams may differ depending on their understanding and implementation of BioC. For a perfect merger and alignment tool, one would need sophisticated algorithms to deal with all possible misalignment scenarios. However, a detailed alert for incompatible BioC files under the current BioC setup may still be informative. Therefore, we refined the BioC Viewer by adding a function that could identify and correct misaligned annotation information. While uploading BioC files, the system checks whether there are files with the same underlying text and format, i.e. same source, date, key and document ID. If such files are detected, the BioC Viewer asks if those files should be merged. If an error occurs during this BioC-importing process, a detailed report is provided.
The BioC Viewer was the front-end for BioGRID curation in the BioCreative V BioC track. Due to the non-competitive nature of the BioC track, the track organizers evaluated the collaborative system by obtaining user feedback from curators, not by using common measures such as precision, recall and F1 scores. In response to design, learnability and usability of the BioC Viewer, BioGRID curators mostly gave positive or strongly positive ratings ( 12 ). In the following sections, we describe the guidelines for the BioC track and the resulting BioC Viewer interface in terms of its functionality.
Methods
Guidelines for the BioC track
Figure 1 presents the workflow of the BioCreative V BioC track. Full-text PMC articles are first annotated by text mining systems. The annotation types include gene/protein/organism mentions and their normalized IDs as well as PPI/GI mention and evidence sentences. The BioC track organizers optimize and unify the text mining results in BioC. These BioC documents are then imported to the visualization tool (BioC Viewer). Finally, BioGRID curators examine the combined results through the viewer, and record PPI/GI pairs using the embedded PPI/GI curation tool. The PPI/GI curation tool is the function that was added on top of the BioC Viewer for BioGRID curation.
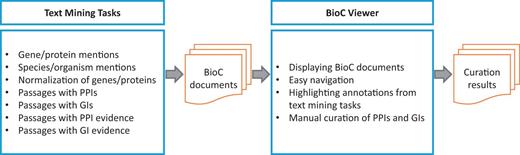
Workflow of the BioCreative V BioC track . Text mining systems first annotate PPI/GI passages as well as gene/protein/organism mentions appearing in full-text articles. After a merging process, BioC documents are imported to the visualization tool (BioC Viewer). Finally, BioGRID curators record PPI/GI pairs by using the BioC Viewer.
The BioC Viewer and the PPI/GI curation tool follow the BioC track guidelines as listed below.
The viewer should provide an easy way to browse annotated text in full-text articles: Users can directly move to a section with annotated passages by clicking section titles in the BioC Viewer.
Some annotations from text mining tasks may overlap each other or occur nested in other annotations. Users should easily recognize these overlaps or nested regions in the viewer: Background colors are used for gene/protein/organism mentions and underlines are used to mark PPI/GI passages.
The BioC Viewer should provide a means to automatically enter normalized gene IDs (i.e. Entrez Gene IDs) to the PPI/GI curation tool if there are any IDs assigned to gene/protein mentions: A mouse click on annotated genes automatically fills the necessary information in the PPI/GI curation tool.
Curators should be able to edit PPI/GI entries if curated gene/protein pairs have errors: Edit icons appear with PPI/GI pairs.
The PPI/GI curation tool supports binary interactions only.
Duplicate PPI/GI pairs may appear when different experimental methods are used for finding the same PPI/GI pair.
An interaction pair is directional. Hence, we may see a PPI/GI with the same genes and the same evidence, but the difference is the order of interacting genes.
Implementation
Our focus for implementing the BioC Viewer was to create a visualization interface that can display annotations from text mining systems. We regularly obtained user feedback from BioGRID curators during the entire development process. Although the viewer did not require a manual annotation function, curators wanted to curate PPI/GI pairs online. Therefore, we added a plug-in function for PPI/GI curation. This is for adding PPI/GI pairs to a database only, and does not associate curated genes with actual positions in full-text articles.
To make a more general BioC viewer, we have added two new functions since the BioCreative V Workshop: ( 1 ) merging multiple BioC files and ( 2 ) correcting misaligned annotations. There are several reasons that can cause annotation misalignment. One case is code points (or characters) versus bytes when computing offsets. For example, using code points, the offset of ‘peptide’ in the phrase, ‘amyloid-β peptide’, is 10. However, when the byte offset is used, the offset of ‘peptide’ in the same phrase becomes 11 in UTF-8. Another case is the existence of more than one version for a document. PubMed and PMC are not static repositories. They are often replaced with new versions, and working with different article versions may cause the misalignment issue. Our solution is straightforward. All offsets are computed based on code points. In any case, if misaligned offsets are identified, the viewer tries to find a match within the same paragraph. One more function we have added to the BioC Viewer is that users can now download the merged files in BioC.
The BioC Viewer uses HTML5/CSS to support most web browsers such as Chrome, Safari, Internet Explorer, Edge and Firefox. The interface was implemented using Ruby on Rails, MySQL, JavaScript and Semantic-UI libraries. It also employs the ‘simple_bioc’ Ruby library ( 3 ) for parsing and building BioC files. The source code of the BioC Viewer is available at https://github.com/dongseop/bioc_viewer .
Results
The BioC Viewer consists of three main pages: list of projects ( Figure 2 ), list of documents ( Figure 3 ) and document viewer ( Figure 4 ). Users can register to use the BioC Viewer with an email address. After a successful login, a user is redirected to the project list page.
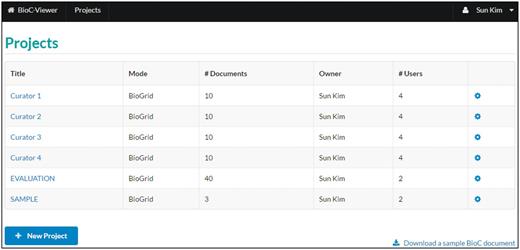
List of projects . A project is a basic unit of the viewer, and each project consists of a set of documents. When a user creates a new project, one of two modes, ‘Normal’ or ‘BioGRID’, can be chosen. In the ‘BioGRID’ mode, the PPI/GI curation tool is visible in the document viewer.
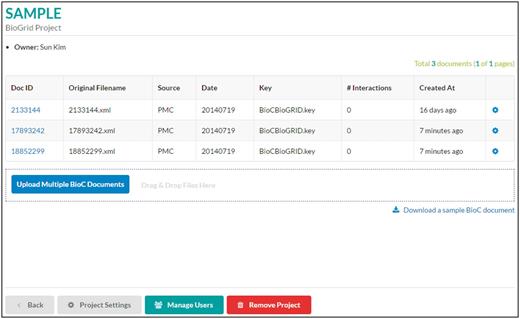
List of documents . A project is a collection of BioC documents. In this page, a user can upload, download or delete BioC documents. Sharing with other users or removing a project can be done using buttons at the bottom of the page.
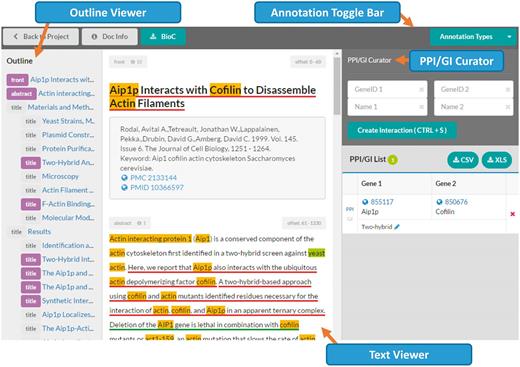
Document viewer in the ‘BioGRID’ mode . The document viewer consists of four main parts: the Annotation Toggle Bar (top), the outline viewer (left side), the text viewer (center) and the PPI/GI curator (right side). The PPI/GI curation tool is only available in the ‘BioGRID’ mode.
List of projects
A project is a basic unit of the BioC Viewer. A user can create an empty project by selecting ‘New Project’. Two modes, ‘Normal’ and ‘BioGRID’, are currently available for a project, and one may switch modes at any time. The difference is in the ‘BioGRID’ mode the PPI/GI curation tool is visible and can be used for PPI/GI curation for BioGRID. The other functions including merging and downloading BioC files are the same for both modes.
Users can manage BioC documents for each project using the setting icon at the end of each row. A project can be shared with collaborators. Through ‘Manage Users’, the owner of a project can assign ‘Read’, ‘Write’ and ‘Admin’ privileges to other users. ‘Read’ assigns users read-only permission for the project; ‘Write’ permission allows users edit privileges for the project. ‘Admin’ permission gives other users the same privileges as the owner, i.e. ‘Admin’ users can add/delete/edit documents and annotations, but also can manage users of the project. Figure 2 shows a logged-in user’s projects. The list may contain the projects that are shared by other users. By clicking the title of a project, a user moves to a ‘list of documents’ page for the project.
List of documents
A project contains a collection of BioC documents as shown in Figure 3 . In this page, a user can upload or delete BioC documents. Users can use ‘Upload Multiple BioC Documents’ or a mouse drag-and-drop for uploading BioC files. While uploading files, the viewer checks whether there are multiple compatible files for the same underlying text document, i.e. files with the same source, date, key and document ID. If such compatible files are found, the interface asks users permission to merge those BioC files. For BioC files that are not compatible with the BioC Viewer, the interface shows an error report of why users cannot import those files. In the current version, we only display and validate annotations. Any relations represented in a BioC document remain untouched, and they are not displayed in the BioC Viewer.
The setting icon at the end of each row can be used for downloading BioC files, exporting PPI/GI curation results or removing documents.
Document viewer
The document viewer consists of four main parts ( Figure 4 ): the annotation toggle bar, the outline viewer, the text viewer, and the PPI/GI curator. Note that the PPI/GI curator is only shown when the ‘BioGRID’ mode is selected.
1) Annotation toggle bar: Using this button, users can make certain annotation types appear or disappear. Different colors can also be selected through this menu. Currently, 12 colors are supported for underlines and background highlights. By default, predefined colors are used for BioGRID annotation types. For other annotation types, the system randomly assigns colors among unused ones. In the same location, the document information (‘Doc Info’) and the BioC download (‘BioC’) buttons are also available.
2) Outline viewer: This provides a bird’s-eye-view of a document. The tool captures section titles of a full-text article, and displays a hierarchy using sentences from titles. Users can jump to a specific section by clicking the section title in the outline viewer. The sections with annotations are marked in orange or purple. For the ‘BioGRID’ mode, purple means the section includes PPI/GI annotations. It is highlighted in orange if a section includes only gene/protein name annotations.
3) Text viewer: This viewer displays a full-text article and annotations. Annotations are highlighted using colors predefined in the annotation toggle bar. Our straightforward approach is that gene/protein/organism mentions use background colors and PPI/GI passages use underlines for highlights. For example, in Figure 4 , the PPI passage is marked in a blue underline. Meanwhile, ‘Aip1p’, ‘Cofilin’, and ‘Actin’ are shown in yellow background color. Due to the restriction of HTML5/CSS, it is difficult to display overlapping or nested annotations. Therefore, the BioC Viewer highlights overlapping annotations by dividing the overlapping region into multiple non-overlapping segments.
4) PPI/GI curator: When the ‘BioGRID’ mode is selected, users can add PPI/GI pairs through this PPI/GI curator. The first part of the curator has a form for creating a new interacting PPI/GI pair. Users can manually enter gene/protein names and their corresponding IDs, but also a mouse click on an annotated gene from the text viewer can fill in the gene name and its assigned ID automatically. Users will be asked for a PPI/GI experimental method after clicking ‘Create Interaction’ or pressing the hotkey, Ctrl + S. Currently, 16 PPI and 11 GI experimental systems are available in the menu. Curated PPI/GI pairs are listed in the bottom of the curator. These are stored in the server once they are created, and can be downloaded in a comma-separated (CSV) or Excel (XLS) file.
Feedback from BioGRID curators
BioCreative competitions have evaluated text-mining performance based on gold-standard sets in general, whereas the BioCreative V BioC track addressed a practical issue by setting up a collaborative task without a gold-standard set. Hence, the performance of the BioC track system was not measured by classification or retrieval metrics, e.g. precision, recall, F1 or average precision. After curating PPI/GI information using the BioC Viewer, four BioGRID curators were asked to rate the usefulness of the system and each functionality on a scale from 1 (bad) to 5 (good) ( 12 ).
The questionnaire as related to the BioC Viewer sought a response to ‘Design of BioC Viewer’, ‘Learning to use BioC Viewer’ and ‘Usability’ ( 12 ). BioGRID curators rated 3.7, 4.3 and 3.5 on average for interface design, learnability and usability, respectively. We obtained 3.0 (neutral) for the question, ‘the interface provided a means to easily correct mistakes’. This is because the interface’s basic function was a viewer, and the curation tool was a plug-in for the viewer. Very limited actions were possible with this setup. However, all curators agreed that the interface was intuitive and easy to use.
Conclusions
The BioC Viewer is a visualization interface for BioC, which was developed to assist BioGRID curators for the BioCreative V BioC track. To meet the requirements of the BioC track, we utilized our implementation of the annotation tool, BioQRator, but extended it to deal with PMC full-text articles. The BioC Viewer was used to display gene/protein/organism mentions and PPI/GI passages. BioGRID curators used the PPI/GI curation tool in the BioC Viewer for curating PPI/GI pairs. Even though the BioC Viewer was initially designed for BioGRID curation, there is no restriction for annotation types, i.e. any other types such as disease and gene ontology annotations can be used.
As a viewer for BioC documents, we realized the importance of correcting annotations misaligned with text and merging multiple compatible document records. Thus, we improved the interface to provide such functionalities. BioC users can now use the BioC Viewer simply to merge different annotations from multiple files, and to confirm BioC compatibility of annotation offsets. By closely working with BioGRID curators and the BioC track organizers, the BioC Viewer received positive feedback in the official evaluation, particularly, in terms of usability and learnability. The BioC Viewer currently supports BioC files with annotation types only. We plan to include relation types in the near future.
Acknowledgements
The authors would like to thank the BioCreative V BioC track organizers and BioGRID curators for their valuable comments, which improved the BioC Viewer.
Funding
The 2007 Research Fund of Myongji University to D.K.; The Genome Program for Fostering New Post-Genome Industry of the National Research Foundation (NRF) funded by the Korean government (MSIP) (NRF-2014M3C9A3064706) to D.K.; The grant (2013-7205) from the Asan Institute for Life Sciences to S.Y.S.; Intramural Research Program of the National Institutes of Health, National Library of Medicine to S.K. and W.J.W.
Conflict of interest. None declared.
References
Author notes
†These authors contributed equally to this work.
Citation details: Shin,S.Y., Kim,S., John Wilbur,W. et al. BioC viewer: a web-based tool for displaying and merging annotations in BioC. Database (2016) Vol. 2016: article ID baw106; doi:10.1093/database/baw106



