-
PDF
- Split View
-
Views
-
Cite
Cite
Justen Manasa, Richard Lessells, Theresa Rossouw, Kevindra Naidu, Cloete Van Vuuren, Dominique Goedhals, Gert van Zyl, Armand Bester, Andrew Skingsley, Katharine Stott, Siva Danaviah, Terusha Chetty, Lavanya Singh, Pravi Moodley, Collins Iwuji, Nuala McGrath, Christopher J. Seebregts, Tulio de Oliveira, Southern African Treatment Resistance Network (SATuRN) RegaDB HIV drug resistance and clinical management database: supporting patient management, surveillance and research in southern Africa, Database, Volume 2014, 2014, bat082, https://doi.org/10.1093/database/bat082
Close - Share Icon Share
Abstract
Substantial amounts of data have been generated from patient management and academic exercises designed to better understand the human immunodeficiency virus (HIV) epidemic and design interventions to control it. A number of specialized databases have been designed to manage huge data sets from HIV cohort, vaccine, host genomic and drug resistance studies. Besides databases from cohort studies, most of the online databases contain limited curated data and are thus sequence repositories. HIV drug resistance has been shown to have a great potential to derail the progress made thus far through antiretroviral therapy. Thus, a lot of resources have been invested in generating drug resistance data for patient management and surveillance purposes. Unfortunately, most of the data currently available relate to subtype B even though >60% of the epidemic is caused by HIV-1 subtype C. A consortium of clinicians, scientists, public health experts and policy markers working in southern Africa came together and formed a network, the Southern African Treatment and Resistance Network (SATuRN), with the aim of increasing curated HIV-1 subtype C and tuberculosis drug resistance data. This article describes the HIV-1 data curation process using the SATuRN Rega database. The data curation is a manual and time-consuming process done by clinical, laboratory and data curation specialists. Access to the highly curated data sets is through applications that are reviewed by the SATuRN executive committee. Examples of research outputs from the analysis of the curated data include trends in the level of transmitted drug resistance in South Africa, analysis of the levels of acquired resistance among patients failing therapy and factors associated with the absence of genotypic evidence of drug resistance among patients failing therapy. All these studies have been important for informing first- and second-line therapy. This database is a free password-protected open source database available on www.bioafrica.net.
Database URL:http://www.bioafrica.net/regadb/
Introduction
The international response to the human immunodeficiency virus (HIV) pandemic has been characterized by unprecedented speed and depth that was only possible through collaboration of clinicians, scientists and civil society including many disciplines and research groups. As a result of this process, the study of the HIV epidemic has generated substantial amount of data. In addition, because HIV is one of the first organisms for which genomic data have been used to identify resistance to drugs and to trace its origin, hundreds of thousands of subgenomic regions have been sequenced both from the management of patients infected by the virus and from academic endeavors to try to understand the epidemic and to discover and develop interventions.
Numerous public databases, such as the Los Alamos HIV Database and the Stanford HIV Drug Resistance Database, have been created to manage the burgeoning number of HIV genomic data sets (1, 2). These databases provide platforms for academics to share and compare data as well as to answer new research questions not originally envisioned by the original investigators. However, a limitation of these public databases is that they do not store and curate data before publication. Moreover, these databases are primarily sequence repositories and contain limited clinical data. Therefore, there is a need for databases to manipulate and curate primary data that include both sequence data and associated clinical, treatment and monitoring data.
In this article, we describe the online database of the Southern African Treatment and Resistance Network (SATuRN). The network is a consortium of clinicians, scientists, public health experts and policy makers (3). The network has 24 member institutions working in southern Africa at the epicenter of the HIV and tuberculosis (TB) epidemics. To foster collaboration among members and to curate primary data, the SATuRN RegaDB HIV Drug Resistance and Clinical Management Database (http://www.bioafrica.net/regadb/index.html) was established. RegaDB is an integrated open source relational database for the management and analysis of HIV treatment, monitoring and resistance data (4). The database is designed to facilitate individual patient management and also to enable real-time surveillance and research, ultimately to inform public health policies in the region.
SATuRN member sites are encouraged to use RegaDB for real-time management of patients failing antiretroviral therapy (ART). It is configured to incorporate a number of online analytic tools such as the Rega HIV Subtyping tool (5) and drug resistance interpretation tools such as HIVDB (6), REGA (7) and ANRS (8, 9). The database is used to produce genotypic resistance reports with specialist advice on therapeutic options tailored to the clinical and treatment history data provided by the clinicians. As a consequence, medical officers and nurses are impelled to provide detailed data to receive robust advice, and this cycle drives quality data curation.
In this article, we first describe the data collection methods followed by a description of the data curation process. We also provide information on database users, data sharing and access policy. We conclude the article with examples of how our curated data have been used for biological discovery.
Data collection and curation process: primary data
A clinical case report form was developed for the collection of demographic and clinical information, treatment data and laboratory monitoring data for input into the SATuRN RegaDB (Supplementary Information). All data are anonymized at the point of entry to the database, but secure records held separately allow linkage to patient care and access to follow-up data. The medical officers or nurses managing the patient send the completed form together with a blood sample submitted for genotypic resistance testing to a central laboratory. The sample is used to produce an HIV-1 DNA sequence of the protease (PR) and reverse transcriptase (RT) genes, which are HIV-1 proteins targeted by first- and second-line ART regimens in southern Africa. Genotypic resistance testing is performed using the in-house SATuRN/Life Technologies method (10). The genotype is translated, and drug resistance mutations are characterized with the use of the drug resistance algorithms contained within RegaDB.
The demographic data collected includes age and sex. Patient identifiers such as names and national identity (ID) numbers are not stored in the RegaDB database. All the stored data are anonymized, and it is the responsibility of the medical officers to link the data to the patient for clinical management. Laboratory data include the patient’s viral load and CD4+ cell count results; in addition, hepatitis B virus surface antigen (HBsAg), creatinine clearance, hemoglobin and alanine aminotransferase results, pertinent to future treatment decisions, are also included.
The treatment information focuses on the ART history as well as treatments for comorbid conditions, in particular TB and hepatitis B virus, as these influence the selection of ART regimens. The start and end dates of specific ART regimens are recorded, with dosages and reasons for any substitution or switch of ART. The clinical form also includes a series of questions relating to adherence, which are based on the assessment tools within the South African national guidelines, and social factors that may influence adherence such as alcohol intake.
Once the HIV genomic data are generated and uploaded, they pass through a quality control step, which includes analysis for deletions, insertions and frame shifts, as well as for contamination using the Basic Local Alignment Search Tool (BLAST) (11) and phylogenetic methods. The genomic sequence is also subtyped using phylogenetic methods that can identify recombinants (5). The PR and RT proteins are analyzed by a pre-selected drug resistance algorithm, such as Stanford HIVDB or REGA algorithm or ANRS, to identify drug resistance mutations and provide an assessment of the level of resistance. The drug resistance interpretation, together with a graphical history of the patient’s ART and laboratory monitoring history, is presented in the form of a report in MS Word (.doc) and rich text format (.rtf) (Figure 1).
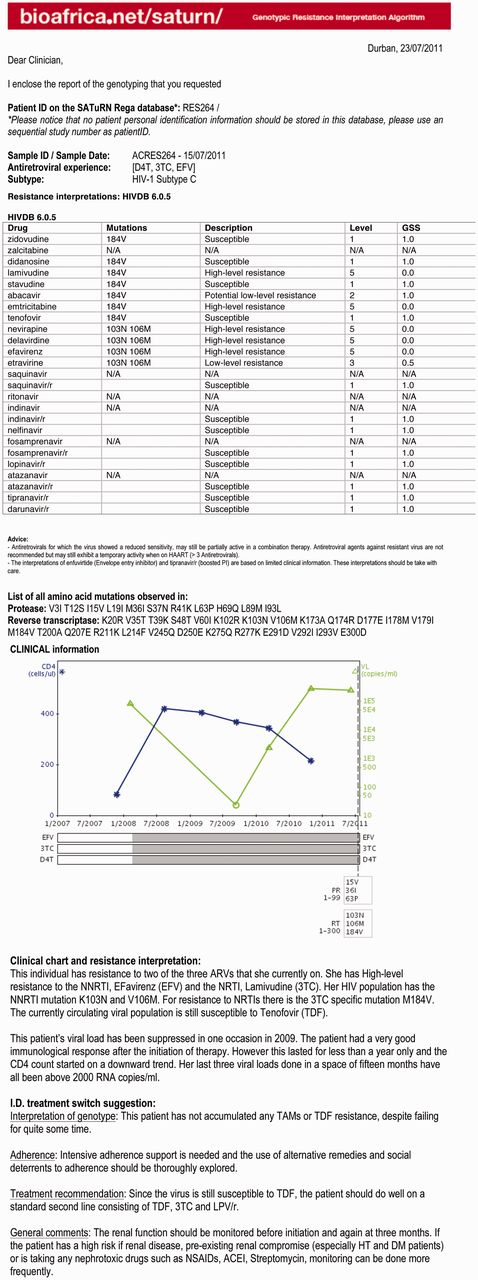
SATuRN Resistance report example. The first page of the report provides a table with drug resistance mutation. The second page contains clinical chart and written interpretation of clinical chart and resistance and a specialized infectious diseases (I.D.) physician switch interpretation.
All SATuRN member institutions are encouraged to use SATuRN’s RegaDB database reports for the management of patients failing ART. Specialist clinician advice in the report adheres, wherever possible, to regimens included within national treatment guidelines. Non-standard regimens are only recommended when there is a strong justification, and any request for ARTs outside the national guidelines requires further approval by the Department of Health. The public health approach to ART is of crucial importance to southern Africa, as >4 million patients are on treatment in the region and the great majority of patients receive treatment at primary health care clinics, with limited pharmaceutical and medical support.
The data are reviewed at many stages to seek out inconsistency and improve quality. Figure 2 shows a model example of the data sources and the review and curation steps. In this model, data from the clinical case report form are added to the database by a team of data curators. Two independent data clerks are involved in the data entry process—one enters the data and the other reviews the data to ensure accurate data entry. In addition, the clinical form is sent to the laboratory staff and the specialist clinicians. The laboratory staff are trained to interpret the clinical chart and to ensure that it makes biological sense (for example, a suppressed viral load result is not usually plausible unless the patient was on treatment at the time). The specialist clinicians ensure that the resistance levels are consistent with the drug regimens received by the patient, and write a detailed therapy recommendation (Figure 1) that is added to the database.
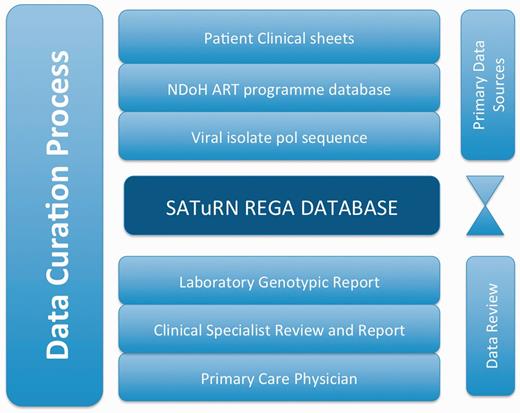
An example of the stages of data curation to increase the quality of the data in the SATuRN RegaDB. The primary data source includes the patient’s clinical information (e.g. viral load, CD4+ count and treatment regimens) that is stored in the patient file, access to National Department of Health ART program database that contains similar clinical information as the patient’s clinical information and the generation of a viral genomic isolate for the pol gene. These data are reviewed by a data curator and added to the database by a data enterer. A report is produced, which is further reviewed by at least one laboratory staff member, a clinical specialist and a physician managing the patient at the primary health care clinic. Any inconsistency on the data is discussed between the individuals reviewing the data and data curator.
The whole process described in Figure 2 takes, on average, 10–14 days. This process is part of the HIV Treatment Failure Clinic (HIV-TFC) model (12). It is important that the data curation is expedited as the patient follow-up visit is normally scheduled for 3–4 weeks after the initial visit at which the blood sample was taken. To receive a detailed report including specialist clinicians comments, the medical officers managing the patients need to complete the clinical form accurately. This motivates them to supply accurate information. Although the processes of data cleaning, validation and curation described here might seem laborious and time-consuming, the quality data obtained from this process are worth the investment. This is especially true in the context of reports of significant weaknesses of the general public health program data in the region (13, 14). Furthermore, the use of a specialized physician to interpret drug resistance results is the norm in South Africa and Botswana public health HIV treatment programs (15). However, a process evaluation has been performed to review the whole system and identify areas that still need further optimization so as to deliver the most cost-efficient system while maintaining quality.
Data collection and curation: published data
Published manuscripts on PubMed with linked sequence data deposited in Genbank, but not yet in the REGA database, are identified on a regular basis to add to the database. The search terms used are ‘HIV-1’, ‘drug resistance’ and ‘Africa’. Only studies with data from patients from southern Africa are included. The manuscripts are reviewed to extract treatment and monitoring data linked to the sequences. If there are limited data, the authors are contacted directly with a request for further information related to the sequences. All published HIV-1 genomic sequences pass through a quality analysis process. These include translation of all sequences and identification of deletions, insertions and frame shifts. Sequences with more than three stop codons, frame shifts, deletions or highly ambiguous nucleotides are flagged and the authors contacted to ensure that these sequence problems are real. Sequences are also checked for duplicates in the RegaDB database with the usage of BLAST and phylogenetic methods. Sequences with high similarity to other sequences in RegaDB (i.e. ≥98%) using a clustering method with bootstrap support (>70%) with another sequence in the phylogeny are flagged as potential duplicates and authors contacted to determine whether these sequences arise from samples from the same individual or whether this is a sequence already deposited at the SATuRN RegaDB.
Database users and data access policies
Currently there are six large cohorts using the SATuRN REGA database for clinical management of patients failing ART (The ANRS 12249 Treatment as Prevention trial in rural KwaZulu-Natal hosted by the Africa Centre for Health and Population Studies, The Hlabisa HIV Treatment and Care Programme in partnership with the Africa Centre for Health and Population Studies, University of Pretoria Medical School, University of Stellenbosch Medical School, University of the Free State Medical School, Inkosi Albert Luthuli Central Hospital, Durban). In addition, another eight groups have published data that have been curated in the REGA database (Table 1). In total, ∼7000 genotypes with related treatment and monitoring data have been collated to date. The data sets increase at a rate of >200 genotypes a month, so the database is continually expanding. Each genotype in the database belongs to a single data set. Genotypes are associated with the previously described attributes (clinical, adherence and demographic information), laboratory measurements (CD4+ cell count, viral load) and ART regimens (start and stop dates) and viral genotypes. Each data set has a unique identifier, access is password-controlled and the owner of the data set has the authority to allocate access to collaborators at different levels (read only, or read and write).
| . | Number of genotypes . | Study participants . | Types of data . |
|---|---|---|---|
| Cohorts, primary data | |||
| Africa Centre for Health and Population Studies, rural KZN, Hlabisa sub-district | 1056 | Adults failing first-line therapy (n = 525), pediatric failing first-/second-line (n = 102), primary resistance (n = 449) | Clinical, treatment, adherence, co-infections |
| University of Pretoria Medical School, Pretoria | 383 | Adults failing first-line therapy (n = 132), pediatric failing first-/second-line (n = 231) | Clinical, treatment, adherence, co-infections |
| University of the Free State, Faculty of Health Sciences, Bloemfontein and surroundings | 874 | Adults failing first-line therapy (n = 538), adults failing second-line (n = 106), pediatrics and adolescents (n = 158) | Clinical, treatment, adherence, co-infections |
| Stellenbosch University, Faculty of Medicine and Health Sciences, Cape Town and surroundings | 1482 | Adults failing first-line ART (n = 601), adults failing second-line (n = 313), pediatric and adolescents (n = 568) | Clinical, treatment |
| Inkosi Albert Luthuli Central Hospital, Durban and surroundings | 115 | Adults failing first-line therapy (n = 90), pediatric failing first/second line (n = 25) | Clinical, treatment |
| ANRS Treatment as Prevention Trial in rural KwaZulu-Natal | 36 | Adults failing first-line therapy (n = 36) | Clinical, treatment, adherence, co-infections |
| Total | 3946 | ||
| Published data | |||
| University of the Free State Medical School (Huang et al.) | 354 | Naive patients before treatment | Clinical tests (VL + CD4) |
| University of Zimbabwe (Dalai et al.) | 210 | Antenatal patients before treatment | Genotype and basic demographic |
| Stellenbosch University, Faculty of Health Sciences, Cape Town and surroundings (ARETAS) (in press) | 341 | Patients failing first-line ART | Clinical, treatment |
| NICD (Seioghe et al.) | 561 | Antenatal patients before and after sdNVP | Genotype and basic demographic |
| AURUM institute (Hoffman et al.) | 167 | Patients failing first-line ART | Clinical, treatment |
| KwaZulu-Natal (Matthews et al., 2008) | 475 | Naive patients before treatment | Genotype and basic demographic |
| NICD (Pillay et al., 2008) | 101 | Antenatal patients before treatment | Genotype and basic demographic |
| Cape Town (Jacobs et al., 2008) | 91 | Naive patients before treatment | Genotype and basic demographic |
| UKZN (Gordon et al., 2003) | 72 | Naive patients before treatment | Genotype and basic demographic |
| Total | 2372 | ||
| . | Number of genotypes . | Study participants . | Types of data . |
|---|---|---|---|
| Cohorts, primary data | |||
| Africa Centre for Health and Population Studies, rural KZN, Hlabisa sub-district | 1056 | Adults failing first-line therapy (n = 525), pediatric failing first-/second-line (n = 102), primary resistance (n = 449) | Clinical, treatment, adherence, co-infections |
| University of Pretoria Medical School, Pretoria | 383 | Adults failing first-line therapy (n = 132), pediatric failing first-/second-line (n = 231) | Clinical, treatment, adherence, co-infections |
| University of the Free State, Faculty of Health Sciences, Bloemfontein and surroundings | 874 | Adults failing first-line therapy (n = 538), adults failing second-line (n = 106), pediatrics and adolescents (n = 158) | Clinical, treatment, adherence, co-infections |
| Stellenbosch University, Faculty of Medicine and Health Sciences, Cape Town and surroundings | 1482 | Adults failing first-line ART (n = 601), adults failing second-line (n = 313), pediatric and adolescents (n = 568) | Clinical, treatment |
| Inkosi Albert Luthuli Central Hospital, Durban and surroundings | 115 | Adults failing first-line therapy (n = 90), pediatric failing first/second line (n = 25) | Clinical, treatment |
| ANRS Treatment as Prevention Trial in rural KwaZulu-Natal | 36 | Adults failing first-line therapy (n = 36) | Clinical, treatment, adherence, co-infections |
| Total | 3946 | ||
| Published data | |||
| University of the Free State Medical School (Huang et al.) | 354 | Naive patients before treatment | Clinical tests (VL + CD4) |
| University of Zimbabwe (Dalai et al.) | 210 | Antenatal patients before treatment | Genotype and basic demographic |
| Stellenbosch University, Faculty of Health Sciences, Cape Town and surroundings (ARETAS) (in press) | 341 | Patients failing first-line ART | Clinical, treatment |
| NICD (Seioghe et al.) | 561 | Antenatal patients before and after sdNVP | Genotype and basic demographic |
| AURUM institute (Hoffman et al.) | 167 | Patients failing first-line ART | Clinical, treatment |
| KwaZulu-Natal (Matthews et al., 2008) | 475 | Naive patients before treatment | Genotype and basic demographic |
| NICD (Pillay et al., 2008) | 101 | Antenatal patients before treatment | Genotype and basic demographic |
| Cape Town (Jacobs et al., 2008) | 91 | Naive patients before treatment | Genotype and basic demographic |
| UKZN (Gordon et al., 2003) | 72 | Naive patients before treatment | Genotype and basic demographic |
| Total | 2372 | ||
| . | Number of genotypes . | Study participants . | Types of data . |
|---|---|---|---|
| Cohorts, primary data | |||
| Africa Centre for Health and Population Studies, rural KZN, Hlabisa sub-district | 1056 | Adults failing first-line therapy (n = 525), pediatric failing first-/second-line (n = 102), primary resistance (n = 449) | Clinical, treatment, adherence, co-infections |
| University of Pretoria Medical School, Pretoria | 383 | Adults failing first-line therapy (n = 132), pediatric failing first-/second-line (n = 231) | Clinical, treatment, adherence, co-infections |
| University of the Free State, Faculty of Health Sciences, Bloemfontein and surroundings | 874 | Adults failing first-line therapy (n = 538), adults failing second-line (n = 106), pediatrics and adolescents (n = 158) | Clinical, treatment, adherence, co-infections |
| Stellenbosch University, Faculty of Medicine and Health Sciences, Cape Town and surroundings | 1482 | Adults failing first-line ART (n = 601), adults failing second-line (n = 313), pediatric and adolescents (n = 568) | Clinical, treatment |
| Inkosi Albert Luthuli Central Hospital, Durban and surroundings | 115 | Adults failing first-line therapy (n = 90), pediatric failing first/second line (n = 25) | Clinical, treatment |
| ANRS Treatment as Prevention Trial in rural KwaZulu-Natal | 36 | Adults failing first-line therapy (n = 36) | Clinical, treatment, adherence, co-infections |
| Total | 3946 | ||
| Published data | |||
| University of the Free State Medical School (Huang et al.) | 354 | Naive patients before treatment | Clinical tests (VL + CD4) |
| University of Zimbabwe (Dalai et al.) | 210 | Antenatal patients before treatment | Genotype and basic demographic |
| Stellenbosch University, Faculty of Health Sciences, Cape Town and surroundings (ARETAS) (in press) | 341 | Patients failing first-line ART | Clinical, treatment |
| NICD (Seioghe et al.) | 561 | Antenatal patients before and after sdNVP | Genotype and basic demographic |
| AURUM institute (Hoffman et al.) | 167 | Patients failing first-line ART | Clinical, treatment |
| KwaZulu-Natal (Matthews et al., 2008) | 475 | Naive patients before treatment | Genotype and basic demographic |
| NICD (Pillay et al., 2008) | 101 | Antenatal patients before treatment | Genotype and basic demographic |
| Cape Town (Jacobs et al., 2008) | 91 | Naive patients before treatment | Genotype and basic demographic |
| UKZN (Gordon et al., 2003) | 72 | Naive patients before treatment | Genotype and basic demographic |
| Total | 2372 | ||
| . | Number of genotypes . | Study participants . | Types of data . |
|---|---|---|---|
| Cohorts, primary data | |||
| Africa Centre for Health and Population Studies, rural KZN, Hlabisa sub-district | 1056 | Adults failing first-line therapy (n = 525), pediatric failing first-/second-line (n = 102), primary resistance (n = 449) | Clinical, treatment, adherence, co-infections |
| University of Pretoria Medical School, Pretoria | 383 | Adults failing first-line therapy (n = 132), pediatric failing first-/second-line (n = 231) | Clinical, treatment, adherence, co-infections |
| University of the Free State, Faculty of Health Sciences, Bloemfontein and surroundings | 874 | Adults failing first-line therapy (n = 538), adults failing second-line (n = 106), pediatrics and adolescents (n = 158) | Clinical, treatment, adherence, co-infections |
| Stellenbosch University, Faculty of Medicine and Health Sciences, Cape Town and surroundings | 1482 | Adults failing first-line ART (n = 601), adults failing second-line (n = 313), pediatric and adolescents (n = 568) | Clinical, treatment |
| Inkosi Albert Luthuli Central Hospital, Durban and surroundings | 115 | Adults failing first-line therapy (n = 90), pediatric failing first/second line (n = 25) | Clinical, treatment |
| ANRS Treatment as Prevention Trial in rural KwaZulu-Natal | 36 | Adults failing first-line therapy (n = 36) | Clinical, treatment, adherence, co-infections |
| Total | 3946 | ||
| Published data | |||
| University of the Free State Medical School (Huang et al.) | 354 | Naive patients before treatment | Clinical tests (VL + CD4) |
| University of Zimbabwe (Dalai et al.) | 210 | Antenatal patients before treatment | Genotype and basic demographic |
| Stellenbosch University, Faculty of Health Sciences, Cape Town and surroundings (ARETAS) (in press) | 341 | Patients failing first-line ART | Clinical, treatment |
| NICD (Seioghe et al.) | 561 | Antenatal patients before and after sdNVP | Genotype and basic demographic |
| AURUM institute (Hoffman et al.) | 167 | Patients failing first-line ART | Clinical, treatment |
| KwaZulu-Natal (Matthews et al., 2008) | 475 | Naive patients before treatment | Genotype and basic demographic |
| NICD (Pillay et al., 2008) | 101 | Antenatal patients before treatment | Genotype and basic demographic |
| Cape Town (Jacobs et al., 2008) | 91 | Naive patients before treatment | Genotype and basic demographic |
| UKZN (Gordon et al., 2003) | 72 | Naive patients before treatment | Genotype and basic demographic |
| Total | 2372 | ||
SATuRN collaborators and external researchers can propose and develop scientific projects and analyses that address new scientific hypotheses. To access comprehensive longitudinal data stored in SATuRN, RegaDB researchers have to submit a project proposal to the SATuRN Executive Committee, which is composed of the SATuRN co-directors, two clinicians and two basic scientist. The project leader must declare his/her intention to develop a project and also declare the main objectives. The project leader should also provide a detailed timeline (e.g. project initiation, data collection and the production of reports, abstracts and manuscripts) and a list of resources available and/or required. A concept sheet is used for the initial submission procedure. The concept sheet and process are described in detail on the SATuRN Web site (http://www.bioafrica.net/saturn).
SATuRN RegaDB genotypes are deposited in GenBank after publication. The data deposited in GenBank are limited to basic demographic information (age and sex), country of origin and isolation year. Genomic data are also deposited in the Stanford HIVDB, complemented with a list of antiretroviral drugs received before the genotype. Furthermore, RegaDB has an automatic export function that is compatible with GenBank and Stanford HIVDB. This is an important process for SATuRN, as one of its aims is to increase the amount of public genotypic drug resistance data in Africa. As part of this process, we have also installed and made publicly available the first mirror of the Stanford HIVDB in Africa (3).
Example of biological discoveries
A literature review and data analysis was performed to review the temporal trends of HIV-1 transmitted drug resistance (TDR) in South Africa (16). Publicly available data were retrieved either from GenBank or by direct request to original authors. Ten data sets with 1618 sequences collected between 2000 and 2010 were pooled, with 72 sequences from recent sero-converters from the Africa Centre’s (AC) 2010 HIV survey in KwaZulu-Natal, South Africa. All of the data were curated and stored in SATuRN RegaDB and were analyzed using the Calibrated Population Resistance Program (17). There was no evidence of TDR from the AC samples.The temporal analysis for South Africa showed that 2002 was the year with the highest TDR rate (6.67, 95% confidence interval: 3.09–13.79%). After 2002, TDR levels decreased to <5% (WHO low-level TDR threshold). There was no statistically significant increase in the interval between 2002 and 2010. These results were published and discussed with the National Department of Health, as they conflicted with a recent publication(18) that pointed to an increase in TDR in KwaZulu-Natal. A large collaborative national survey involving SATuRN investigators and the National Department of Health is currently in process. Continuous representative TDR surveys are needed to ensure that current first-line regimens remain effective, as an increase in TDR could reverse the gains of ARV rollout.
Results from an analysis of data on SATuRN RegaDB for patients with first-line antiretroviral treatment failure in a rural primary health care program in KwaZulu-Natal were recently published (19). A secondary analysis to identify factors associated with the absence of HIV drug resistance in patients with failure of first-line ART to inform adherence strategies and to determine whether unnecessary switches to second-line therapy could be avoided was also done (20). In total, 243 patients were included in the final analysis, and detailed adherence and clinical information was curated from clinics in the Hlabisa Treatment and Care Programme, South Africa (Appendix). The genotypes were linked to 38 other adherence and clinical variables. This information was reviewed by two data clerks and a medical officer and added to RegaDB. Predictors associated with the absence of drug resistance were analyzed by univariable and multivariable logistic regression methods. These data curation and analysis showed that there are a number of factors associated with the absence of drug resistance following ART failure, one of which (baseline CD4+ count) is a strong association with poor adherence, which can lead to significantly higher levels of immunological failure, putting these patients at increased risk of mortality.
The data stored in SATuRN RegaDB are currently being used in many research projects. This article also serves to provide details of existence of these longitudinal data sets that are available for applications from researchers to analyze the data, as described in the SATuRN Manual of Operation. In addition, the HIV drug resistance data curated in the SATuRN RegaDB have provided the information and tools to enable the education and training of health care workers and patients. At the time of writing this publication, clinical cases from the database have been presented to ∼2050 medical officers and nurses throughout Africa at the annual SATuRN conferences and workshops. Moreover, 15 cases stored within RegaDB, which highlight some of the major challenges involved in managing patients failing ART in the public sector in southern Africa, were collated and published recently as an open access book (21), of which ∼10 000 copies were freely distributed by SATuRN, Keth’impilo, Medicine San Fronteirs (MSF) and the Southern African HIV Clinicians Society to their medical officer members.
Conclusion
The SATuRN RegaDB presented in this article is a useful tool for patient management, data curation and capacity building. The combination of curated HIV sequence, clinical and demographic data in this database has been used to identify important factors that are associated with drug resistance in ART-naïve and ART-experienced patients in southern Africa. The SATuRN RegaDB has the potential to become a useful resource for the regional and international research community, similar to the United Kingdom (22) and Swiss National Drug Resistance Databases (23, 24). The data and database will be used to inform local and national policy makers on the level of HIV-1 drug resistance in southern Africa. Furthermore, guidelines of the Southern African HIV Clinicians Society (25) have suggested that the SATuRN drug resistance databases be used as a national model for the management of patients failing ART. The South African National Department of Health and the Botswana Ministry of Health are in the process of piloting the HIV-TFC system. In addition, efforts are currently being made to have more contributions from other southern African researchers as well as to reach out to other countries in sub-Saharan Africa as a whole.
Acknowledgements
The authors thank the data curators from the Africa Centre for Health and Population Studies: Lungani Ndwandwe, Xolile Xineri, Zakhona Gumede; the data capturers at the University of the Free State; Stellenbosch University: M. Seleka and R. Taylor. They also thank the nurses and counselors of the Hlabisa HIV Treatment and Care Program, South Africa, and the laboratory staff of the Africa Centre for Health and Population Studies: Surenshe Pillay; University of the Free State.
Funding
This implementation of resistance testing was supported by the Delegation of the European Union to South Africa (SANTE 2007 147–790; drug resistance surveillance and treatment monitoring network for the public sector HIV antiretroviral treatment program in the Free State), the US Centre for Diseases Control via CAPRISA [project title: Health Systems Strengthening and HIV Treatment Failure (HIV-TFC)]. R.J.L. and N.Mc.G. are supported by the Wellcome Trust [grants 090999/Z/09/Z and WT083495MA, respectively]. The Africa Centre receives core funding from the Wellcome Trust [082384/Z/07/Z]. The Hlabisa HIV Treatment and Care Programme has received support through the United States Agency for International Development (USAID) and the President’s Emergency Plan (PEPFAR) under the terms of award number 674-A-00-08-00001-00. The opinions expressed herein are those of the authors and do not necessarily reflect the view of the USAID or the United States Government. None of the funders had a role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding for open access charge: Wellcome Trust.
Conflict of interest. None declared.
References
Author notes
Citation details: Manasa,J., Lessells,R., Rossouw,T. et al. Southern African Treatment Resistance Network (SATuRN) regaDB HIV drug resistance and clinical management database: supporting patient management, surveillance and research in southern Africa. Database (2013) Vol. 2013: article ID bat082; doi:10.1093/database/bat082



