-
PDF
- Split View
-
Views
-
Cite
Cite
Shuai Yang, Xingwei Zhang, Huayang Li, Yudong Chen, Long Yang, GAN: a platform of genomics and genetics analysis and application in Nicotiana, Database, Volume 2018, 2018, bay001, https://doi.org/10.1093/database/bay001
Close - Share Icon Share
Abstract
Nicotiana is an important Solanaceae genus, and plays a significant role in modern biological research. Massive Nicotiana biological data have emerged from in-depth genomics and genetics studies. From big data to big discovery, large-scale analysis and application with new platforms is critical. Based on data accumulation, a comprehensive platform of Genomics and Genetics Analysis and Application in Nicotiana (GAN) has been developed, and is publicly available at http://biodb.sdau.edu.cn/gan/. GAN consists of four main sections: (i) Sources, a total of 5267 germplasm lines, along with detailed descriptions of associated characteristics, are all available on the Germplasm page, which can be queried using eight different inquiry modes. Seven fully sequenced species with accompanying sequences and detailed genomic annotation are available on the Genomics page. (ii) Genetics, detailed descriptions of 10 genetic linkage maps, constructed by different parents, 2239 KEGG metabolic pathway maps and 209 945 gene families across all catalogued genes, along with two co-linearity maps combining N. tabacum with available tomato and potato linkage maps are available here. Furthermore, 3 963 119 genome-SSRs, 10 621 016 SNPs, 12 388 PIPs and 102 895 reverse transcription-polymerase chain reaction primers, are all available to be used and searched on the Markers page. (iii) Tools, the genome browser JBrowse and five useful online bioinformatics softwares, Blast, Primer3, SSR-detect, Nucl-Protein and E-PCR, are provided on the JBrowse and Tools pages. (iv) Auxiliary, all the datasets are shown on a Statistics page, and are available for download on a Download page. In addition, the user’s manual is provided on a Manual page in English and Chinese languages. GAN provides a user-friendly Web interface for searching, browsing and downloading the genomics and genetics datasets in Nicotiana. As far as we can ascertain, GAN is the most comprehensive source of bio-data available, and the most applicable resource for breeding, gene mapping, gene cloning, the study of the origin and evolution of polyploidy, and related studies in Nicotiana.
Database URL: http://biodb.sdau.edu.cn/gan/
Introduction
The genus Nicotiana is the fourth largest genus in the family Solanaceae. Extensive research has accumulated a huge amount of data regarding genetics, evolution, genomics, taxonomy and breeding in the genus (1). Tobacco (Nicotiana tabacum L.) is cultivated worldwide as a plant of economic importance, as well as a model system in plant biotechnology (2). The collection and utilization of germplasm are the foundation of botanical research. A generally accepted international standard taxonomy for Nicotiana has existed for decades. It consists of 3 sub-genera, 14 sections and 66 species (3). However, most researchers now tend to classify Nicotiana into 13 sections and 76 species (4, 5). Regardless, several Nicotiana germplasm storehouses have been built over the years to collect and maintain Nicotiana germplasm resources. Presently, approximately 2152 Nicotiana accessions are maintained at the NPGS (National Plant Germplasm System, a world-famous genetic diversity warehouse for N.tabacum, http://www.ars-grin.gov/npgs/index.html) (6). In addition, a total of 1160 Nicotiana accessions are held at the CGRIS (Chinese Crop Germplasm Resources Information System, http://icgr.caas.net.cn/cgris_english.html) (7). Furthermore, the Tobacco Genetics and Breeding database (TGB, http://biodb.sdau.edu.cn/tgb/) also provides 1472 Nicotiana accessions with accompanying detailed annotations (8). Genus Nicotiana germplasm is generally understood to be available under >5000 accessions worldwide; however, no systematic database exists to preserve and catalogue all these germplasm resources.
The N. tabacum genome remained a challenge to sequence for quite a long time, because it is allotetraploid and very large. The allotetraploid (2n = 4x = 48) N. tabacum genome contains 24 pairs of chromosomes, derived from the two diploid species N. sylvestris and N. tomentosiformis (9, 10). High-throughput sequencing technologies have developed rapidly; comparative genomics can now provide insights into seven Nicotiana species: the allotetraploid N. benthamiana (11); three diploid species, N. otophora, N. sylvestris and N. tomentosiformis (10) and three common cultivated species, N. tabacum BX, N. tabacum K326 and N. tabacum TN90 (12). In the recent years, there was numerous sequences read archives available for other Nicotiana species like N. obtusifolia, N. nudicaulis and N. repanda and so on. Some of them were successfully used to identify allopolyploid species origin at a great accuracy (13, 14). So the high research value promoted to construct a genome information database which would provide convenient access to all the data.
The establishment of molecular markers and genetic linkage maps has greatly influenced gene mapping, gene cloning, quantitative trait loci and marker-assisted selection research (15, 16). As a model plant, genetic analyses in different Nicotiana genera employ a diversity of molecular markers (17–20). Molecular markers, restriction fragment length polymorphisms (RFLPs) (21), random amplified polymorphic DNAs (RAPDs) (22), amplified fragment length polymorphisms (AFLPs) (23), short sequence repeats (SSRs) (24), intron length polymorphisms (25), single nucleotide polymorphisms (SNPs) (26) and inter-simple sequence repeats (ISSRs) (27), were all excavated from previous research on a large scale. Ten Nicotiana genetic linkage maps were developed based on these markers, in our previous studies (28). However, these results were all derived from contig or scaffold sequences, not from whole genomes. Consequently, these marker linkage analyses needed to be redone on a large-scale using genome sequences.
An excellent resource for the online tobacco research community, the TGB database contains 1472 Nicotiana germplasms, with accompanying detailed annotations and 12 388 potential intron polymorphisms (PIPs), 10 551 EST-simple sequence repeats and 66 297 genomic-SSR markers (G-SSRs) (8). However, with the huge amount of new bio-data emerging in recent years, TGB can not satisfy all new research needs. Therefore, we found it necessary to establish a comprehensive platform of Genomics and Genetics Analysis and Application in Nicotiana (GAN, http://biodb.sdau.edu.cn/gan/). GAN contains a Source Section (Germplasm and Genomes), a Genetics Section (Markers, Genetics Maps and Genetics Annotations), a Tools Section [JBrowse (29) and Online Tools] and an Auxiliary Section (Datasets, Download and Manual) for Nicotiana research. GAN will greatly expedite genomics and genetics research in Nicotiana and related plants.
Platform content and web interface
GAN provides information regarding the detailed annotation of 5267 Nicotiana accessions, 7 genomes, 10 genetic linkage maps, 2239 KEGG metabolic pathway maps, 209 945 gene families, two co-linearity maps, 3 963 119 genome-SSRs, 10 621 016 SNPs, 12 388 PIPs, 102 895 reverse transcription-polymerase chain reaction (RT-PCR) primers, JBrowse and 5 useful online softwares. The GAN Web interface was designed to include the following components: Source, Genetics, Tools and Auxiliary (Figure 1).
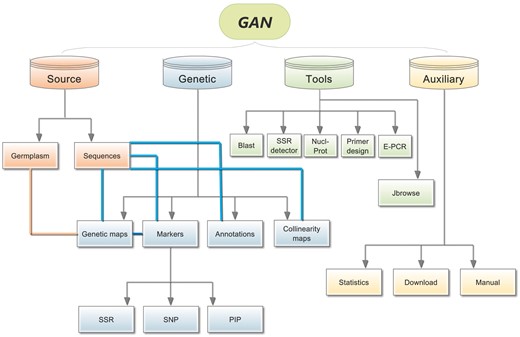
Overall GAN framework. The four different colours represent the four different main sections of GAN, Source, Genetics, Tools. Connections are illustrated between related subsections using straight lines with different colours.
JBrowse and the five online bioinformatics tools, Blast (30), Primer3 (31), SSR-detect (32), Nucl-Protein and E-PCR (33), are provided on the JBrowse and Tools pages. An Auxiliary Section contains all the datasets in GAN, displayed on a Statistics page and available for download on the Download page. In addition, the Auxiliary Section contains a user’s manual, provided on the Manual page in English and Chinese languages. GAN provides a user-friendly Web interface for searching, browsing and downloading genomics and genetics datasets for Nicotiana. As far as we know, GAN provides the most comprehensive Nicotiana bio-data available, and is an excellent resource for research in Nicotiana breeding, gene mapping, gene cloning, the origin and evolution of polyploidy and related studies.
The GAN platform provides researchers all necessary data, and is composed of five main parts: Germplasm, Genomics, Genetics, Markers and Tools (Figure 2). (i) The Germplasm Section holds 5267 Nicotiana accessions, which can be queried using several different routes: specific ID, species, Nicotiana type, cultivar origin, agronomic characteristics and disease resistance characteristics. Furthermore, detailed germplasm information is also provided with high-quality images from the field or laboratory. (ii) The Genomics Section contains the seven available Nicotiana genome sequences, and all sources and annotations. (iii) The Genetics Section contains genetic maps, gene annotations, synteny analyses and gene family make up analyses for those seven Nicotiana species with complete genome data. (iv) The Markers Section contains all of the three widely used molecular markers, SSRs, SNPs and PIPs, that have been detected in those seven Nicotiana genes, and some of the exons, introns and UTRs. Furthermore, because RT-PCR is such a sensitive and widely used method for the detection of mRNA expression levels, RT-PCR primers are also available, predesigned using the coding sequences (CDSs) of current Nicotiana species. (v) The Tools Section contains the online bioinformatics tools Blast, SSR-detect, Primer3, Nucl-Protein and E-PCR, which have all been integrated into GAN, as well as the currently popular genome browser JBrowse for the display of detailed annotation and marker information overlaid on the genome.

Main GAN Web page. (A) GAN homepage: provides quick entry paths to all main parts. (B) Germplasm page: stores 5267 Nicotiana accessions, detailed information and a seven-mode search mechanism. (C) Genetics page: consists of genetic maps, gene annotations, synteny analyses, gene family identifications and sequence query. (D) Markers page: includes all SSR, SNP, PIP and RT-PCR markers in GAN. (E and F) Tools and JBrowse pages: allow access to online bioinformatics tools and the genome browser JBrowse for detailed information overlays and analyses.
Germplasm
We divide the 5267 GAN accessions into 35 Nicotiana species, of which ∼92.8% are N. tabacum L., in accordance with accepted Nicotiana classification. The germplasm type is differentiated into seven tobacco categories: Aromatic, Burly, Cigar, Flue-cured, Wild, Rustica and Sun-cured. The 5267 Nicotiana accessions come from 45 different cultivar origins, with most cultivar originating in China and the next most from North America (Table 1). Detailed disease resistance information is provided for all accessions, including Black Shank, Granville Wilt, Root-knot Nematode, Brown Spot, Tobacco mosaic virus, Cucumber mosaic virus and Potato virus Y (Figure 3).
| Cultivar Origin . | Aromatic tobacco . | Burley tobacco . | Cigar tobacco . | Flue-cured tobacco . | Wild tobacco . | Rustica tobacco . | Sun-curing tobacco . | Total . |
|---|---|---|---|---|---|---|---|---|
| China | 17 | 90 | 5 | 1762 | 0 | 328 | 2270 | 4472 |
| America | 0 | 64 | 13 | 287 | 4 | 5 | 27 | 400 |
| Zimbabwe | 3 | 3 | 0 | 17 | 0 | 1 | 0 | 24 |
| Poland | 4 | 4 | 6 | 5 | 0 | 0 | 2 | 21 |
| Yugoslavia | 8 | 0 | 3 | 4 | 0 | 0 | 2 | 17 |
| Japan | 6 | 4 | 0 | 13 | 1 | 3 | 4 | 31 |
| Bulgaria | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| Canada | 0 | 0 | 0 | 12 | 0 | 0 | 1 | 13 |
| Australia | 0 | 0 | 0 | 3 | 6 | 0 | 1 | 10 |
| Others | 43 | 35 | 28 | 100 | 24 | 7 | 28 | 265 |
| Total | 95 | 200 | 55 | 2203 | 35 | 344 | 2335 | 5267 |
| Cultivar Origin . | Aromatic tobacco . | Burley tobacco . | Cigar tobacco . | Flue-cured tobacco . | Wild tobacco . | Rustica tobacco . | Sun-curing tobacco . | Total . |
|---|---|---|---|---|---|---|---|---|
| China | 17 | 90 | 5 | 1762 | 0 | 328 | 2270 | 4472 |
| America | 0 | 64 | 13 | 287 | 4 | 5 | 27 | 400 |
| Zimbabwe | 3 | 3 | 0 | 17 | 0 | 1 | 0 | 24 |
| Poland | 4 | 4 | 6 | 5 | 0 | 0 | 2 | 21 |
| Yugoslavia | 8 | 0 | 3 | 4 | 0 | 0 | 2 | 17 |
| Japan | 6 | 4 | 0 | 13 | 1 | 3 | 4 | 31 |
| Bulgaria | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| Canada | 0 | 0 | 0 | 12 | 0 | 0 | 1 | 13 |
| Australia | 0 | 0 | 0 | 3 | 6 | 0 | 1 | 10 |
| Others | 43 | 35 | 28 | 100 | 24 | 7 | 28 | 265 |
| Total | 95 | 200 | 55 | 2203 | 35 | 344 | 2335 | 5267 |
| Cultivar Origin . | Aromatic tobacco . | Burley tobacco . | Cigar tobacco . | Flue-cured tobacco . | Wild tobacco . | Rustica tobacco . | Sun-curing tobacco . | Total . |
|---|---|---|---|---|---|---|---|---|
| China | 17 | 90 | 5 | 1762 | 0 | 328 | 2270 | 4472 |
| America | 0 | 64 | 13 | 287 | 4 | 5 | 27 | 400 |
| Zimbabwe | 3 | 3 | 0 | 17 | 0 | 1 | 0 | 24 |
| Poland | 4 | 4 | 6 | 5 | 0 | 0 | 2 | 21 |
| Yugoslavia | 8 | 0 | 3 | 4 | 0 | 0 | 2 | 17 |
| Japan | 6 | 4 | 0 | 13 | 1 | 3 | 4 | 31 |
| Bulgaria | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| Canada | 0 | 0 | 0 | 12 | 0 | 0 | 1 | 13 |
| Australia | 0 | 0 | 0 | 3 | 6 | 0 | 1 | 10 |
| Others | 43 | 35 | 28 | 100 | 24 | 7 | 28 | 265 |
| Total | 95 | 200 | 55 | 2203 | 35 | 344 | 2335 | 5267 |
| Cultivar Origin . | Aromatic tobacco . | Burley tobacco . | Cigar tobacco . | Flue-cured tobacco . | Wild tobacco . | Rustica tobacco . | Sun-curing tobacco . | Total . |
|---|---|---|---|---|---|---|---|---|
| China | 17 | 90 | 5 | 1762 | 0 | 328 | 2270 | 4472 |
| America | 0 | 64 | 13 | 287 | 4 | 5 | 27 | 400 |
| Zimbabwe | 3 | 3 | 0 | 17 | 0 | 1 | 0 | 24 |
| Poland | 4 | 4 | 6 | 5 | 0 | 0 | 2 | 21 |
| Yugoslavia | 8 | 0 | 3 | 4 | 0 | 0 | 2 | 17 |
| Japan | 6 | 4 | 0 | 13 | 1 | 3 | 4 | 31 |
| Bulgaria | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| Canada | 0 | 0 | 0 | 12 | 0 | 0 | 1 | 13 |
| Australia | 0 | 0 | 0 | 3 | 6 | 0 | 1 | 10 |
| Others | 43 | 35 | 28 | 100 | 24 | 7 | 28 | 265 |
| Total | 95 | 200 | 55 | 2203 | 35 | 344 | 2335 | 5267 |
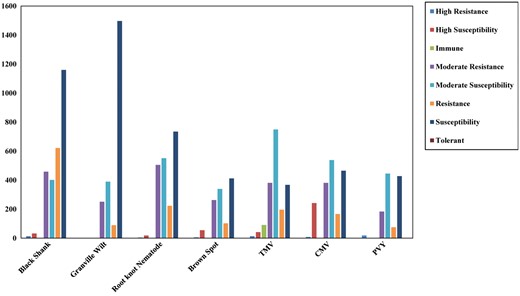
Detailed disease resistance information for the GAN Nicotiana accessions.
The Germplasm page can be searched using ID-Name, species, type, cultivar origin, agronomic characteristics and disease resistance keywords. Inputting your desired words and clicking the ‘Keywords Search’ button initiates a global keyword search. Users can also click the ‘Submit’ button in the Germplasm gray-box to browse all germplasms, displaying all 35 Nicotiana species, 7 Nicotiana types and 45 cultivar origins, which can then be chosen among. Finally, three different specific identifiers, the germplasm’s name, its GAN ID and its National Uniform Number, can be used to search for a specific germplasm. Thirty-five common field agronomic characteristics are provided including phenotype and physiology. Users can choose one or more parameters to generate exact results. Similar to agronomic characteristics, the disease resistance search box offers seven kinds of disease, including different levels of resistance, such that the user can obtain accurate germplasm identifications. Results in the Web interface appear as simple information entries in tables in which users can click a ‘Detail’ hyperlink to generate detailed information about a specific germplasm. The Detail Web page displays four main attributes: basic information, agronomic characteristics, disease resistance and Nicotiana images for each germplasm. The basic information entry provides the Nicotiana species of the germplasm along with an introduction, common names, economic importance and references.
Genomics
Seven sequenced Nicotiana genomes, including assembly and annotation information, genome size (∼2.7 G through ∼5.18 G), contigs and scaffolds can be found on the Genomics page.
Genetics
Ten specific genetic maps were constructed in previous research using different RFLP, RAPD, ISSR and SSR molecular markers. Wild species, Burly and Flue-cured are all used as materials among the 24 chromosomes. Enzymes of 2115 relate to metabolic pathways found in 6 Nicotiana CDSs, which participate in 383 specific pathway maps. A total of 506 553 protein domains and/or families were identified using Pfam, Pkinase, WD domain and PPR family analyses in the 6 Nicotiana CDSs.
Users can browse ten genetics maps in the Genetics Section. Tables store constitutive data for these genetic maps in the main Web page. A graphical ‘Go’ button is pressed to get the requested genetic map; the maps contain detailed marker information obtained by clicking on the marker. KEGG annotation can then be queried if users select a specific Nicotiana species and input a sequence ID or KEGG Orthology number. Gene family, Nicotiana species or type, Gene ID, PF(Plant Family) accession and name can all be used to start the search.
Molecular markers
A total of 3 429 801 G-SSRs, 326 253 gene-SSRs and 18 485 CDS-SSRs were catalogued in this research. Additionally, 267 699 primer pairs were designed for Nicotiana genes. SNP markers of 10 621 016 were searched against and located on the reference N. benthamiana genome (Table 2). Furthermore, 12 338 PIP makers were obtained from the TGB platform. Finally, 102 859 pairs of E-PCR verified primers were generated for scientific research.
| Species . | Genome-SSR . | Gene-SSR . | SNP . |
|---|---|---|---|
| Nicotiana benthamiana | 492 540 | 61 568 | - |
| Nicotiana otophora | 358 420 | - | 785 204 |
| Nicotiana sylvestris | 420 177 | 63 159 | 1 872 257 |
| Nicotiana tabacum BX | 594 364 | 47 536 | 2 391 165 |
| Nicotiana tabacum K326 | 620 818 | 45 437 | 2 403 132 |
| Nicotiana tabacum TN90 | 637 331 | 44 660 | 2 442 813 |
| Nicotiana tomentosiformis | 306 151 | 63 893 | 726 445 |
| Species . | Genome-SSR . | Gene-SSR . | SNP . |
|---|---|---|---|
| Nicotiana benthamiana | 492 540 | 61 568 | - |
| Nicotiana otophora | 358 420 | - | 785 204 |
| Nicotiana sylvestris | 420 177 | 63 159 | 1 872 257 |
| Nicotiana tabacum BX | 594 364 | 47 536 | 2 391 165 |
| Nicotiana tabacum K326 | 620 818 | 45 437 | 2 403 132 |
| Nicotiana tabacum TN90 | 637 331 | 44 660 | 2 442 813 |
| Nicotiana tomentosiformis | 306 151 | 63 893 | 726 445 |
| Species . | Genome-SSR . | Gene-SSR . | SNP . |
|---|---|---|---|
| Nicotiana benthamiana | 492 540 | 61 568 | - |
| Nicotiana otophora | 358 420 | - | 785 204 |
| Nicotiana sylvestris | 420 177 | 63 159 | 1 872 257 |
| Nicotiana tabacum BX | 594 364 | 47 536 | 2 391 165 |
| Nicotiana tabacum K326 | 620 818 | 45 437 | 2 403 132 |
| Nicotiana tabacum TN90 | 637 331 | 44 660 | 2 442 813 |
| Nicotiana tomentosiformis | 306 151 | 63 893 | 726 445 |
| Species . | Genome-SSR . | Gene-SSR . | SNP . |
|---|---|---|---|
| Nicotiana benthamiana | 492 540 | 61 568 | - |
| Nicotiana otophora | 358 420 | - | 785 204 |
| Nicotiana sylvestris | 420 177 | 63 159 | 1 872 257 |
| Nicotiana tabacum BX | 594 364 | 47 536 | 2 391 165 |
| Nicotiana tabacum K326 | 620 818 | 45 437 | 2 403 132 |
| Nicotiana tabacum TN90 | 637 331 | 44 660 | 2 442 813 |
| Nicotiana tomentosiformis | 306 151 | 63 893 | 726 445 |
Generally, all of the Markers search pages require a Nicotiana species to be chosen as a necessary option. Different markers have their own search options beyond that: The SSR page provides options for sequence type, Gene ID, SSR motif and repeat number; the SNP page allows for searching by genome sequence ID, reference base and query base; the PIP page contains options for PIP ID, PlantGDB ID(http://www.plantgdb.org/) and gene name; the RT-PCR page requires a gene sequence ID. Clicking the ‘Detail’ hyperlink in the Result Web interface for all markers displays detailed information on the marker.
Tools
Five frequently used software tools are provided in GAN (Blast, Primer3, SSR-detect, Nucl-Protein and E-PCR) and one genome browser (JBrowse). Generally speaking users can paste sequences in FastA format directly into a text-area, or upload sequence files from a local computer, into the five software pages. Users select from five Blast variants (blastp, blastx, blastn, tblastx and tblastn) on the Blast page, and search against any of the seven Nicotiana species genome sequences (or corresponding translations) along with basic program parameters to obtain results. In the SSR-detector page, SSRs can be identified using the default parameters of MISA (34). The Nucl-Protein page provides a tool for the translation of nucleotide sequences to protein sequences; users only need to provide a nucleotide sequence and working name, similarly with SSR-detector. The Primer Design page requires users to input the Min-TM, Max-TM, Min-GC, Max-GC and return number to predict maximal Primer3 results. The primer verification tool E-PCR uses the Primer3 results and requires users to select one Nicotiana species as a template, followed by a few alternative parameters. Results are obtained by online perusal or can be download to the local computer.
JBrowse is an open-access genome browser that allows for the storage and display of genome sequences, genes, mRNAs, CDSs, introns, exons, UTRs, SSRs and SNPs, providing detailed annotation for each. In our implementation, the left column lists options consisting of Nicotiana genomes, a GFF choice, SNP loci, SNP coverage and SSR locations. The right column will display the corresponding information when users click the square box before the options in the left column. The GFF option provides an example showing how to work with the bowser. By clicking the GFF square all genome information will graphically display, then a dialog box will pop up if you click a specific graphic. Users can browse and download detailed information from the dialog box. We have added several new and useful features to the basic JBrowse browser (Figure 2).
Download and help
Users can download all the data used in GAN on the Download page. A download tree is used to access all the data relative to the Germplasm, Genomics, Genetics and Markers pages.
Conclusions and perspectives
GAN is a platform for providing Nicotiana accessions while integrating useful genomic and genetic data, and supplying relevant annotations and molecular markers in Nicotiana. The 5267 Nicotiana accessions are the main content of the platform; however, GAN also expedites scientific work by identifying Nicotiana gene annotations and markers and primers for genetics analysis and breeding. Furthermore, the incorporation of several useful tools and JBrowse make it easy for users unfamiliar with bioinformatics to perform big-data scientific research. The platform will be updated continuously as new Nicotiana accessions are collected, and new Nicotiana genome sequences are generated.
Materials and methods
Germplasm and sequences
Information on 51 traits, and plant and inflorescence images of 5267 Nicotiana accessions were exhaustively collected. The genome sequences of N. benthamiana, N. otophora, N. tabacum BX, N. tabacum K326 and N. tabacum TN90 were downloaded from SGN (35) (Sol Genomics Network, https://solgenomics.net/) and the genome sequences of N. sylvestris and N. tomentosiformis were obtained from NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/).
Genetic maps and annotation
Ten Nicotiana genetic linkage maps with associated molecular markers were collected from previous research. Co-linearity maps with tomato and potato were also extracted from previous Nicotiana sequence research (12). All relevant information can be found on the appropriate GAN page.
The KAAS Web interface (36) (http://www.genome.jp/tools/kaas/) was used annotate data with the KEGG pathway assignments (37) (Kyoto Encyclopedia of Genes and Genomes). PfamScan software (38) (http://www.ebi.ac.uk/Tools/pfa/pfamscan/) was used to scan gene families from Pfam (39) (http://pfam.xfam.org/).
Marker identification
From different type sequences of Nicotiana species, the SSRs (Genome-SSR, Gene-SSR, Cds-SSR, Exon-SSR and Intron-SSR) were detected by scanning monomer, dimer, trimer, tetramer, pentamer and hexamer nucleotide motifs with at least 10, 6, 5, 5, 5 and 5 repeats by using perl-based MISA program (http://pgrc.ipk-gatersleben.de/misa/), respectively. For complex SSRs, the maximum difference between two SSRs had to be <100 bp. Primer3 was used for SSR primers design (31). Using N. benthamiana sequences as reference, and the other six Nicotiana species sequences as queries, SNPs were extracted and filtered using the BWA (Burrows-Wheeler Aligner) (40), Samtools (41) and Bcftools. Among them, BWA is responsible for database building and comparison. Samtools is responsible for processing and converting formats. Finaly, Bcftools was used for SNP calling. All parameters were used at default setings.
Platform implementation
The LAMP framework (Linux, Apache, MySQL and PHP/Perl) was used to construct the GAN platform. All standardized data were imported into MySQL for storage and management. Commands are submitted in the Web-friendly interface using HTML on the Apache Web server, generating PHP scripts, which extract corresponding data from MySQL. Perl and Java scripts are used to enhance the appearance of the Web interface and to improve user query efficiency.
Acknowledgement
This work was supported by the Foundation of Shandong Province Modern Agricultural Technology System Innovation Team from the Shandong Agricultural University (No. SDAIT-25-02).
Conflict of interest. None declared.
References
Author notes
Citation details: Yang,S., Zhang,X., Li,H. et al. GAN: a platform of genomics and genetics analysis and application in Nicotiana. Database (2018) Vol. 2018: article ID bay001; doi:10.1093/database/bay001
Shuai Yang and Xingwei Zhang contributed equally to this work.



