-
PDF
- Split View
-
Views
-
Cite
Cite
A Groenewoud, G Forn-Cuní, F B Engel, B E Snaar-Jagalska, XePhIR: the zebrafish xenograft phenotype interactive repository, Database, Volume 2022, 2022, baac028, https://doi.org/10.1093/database/baac028
Close - Share Icon Share
Abstract
Zebrafish xenografts are an established model in cancer biology, with a steadily rising number of models and users. However, as of yet, there is no platform dedicated to standardizing protocols and sharing data regarding zebrafish xenograft phenotypes. Here, we present the Xenograft Phenotype Interactive Repository (XePhIR, https://www.xephir.org) as an independent data-sharing platform to deposit, share and repurpose zebrafish xenograft data. Deposition of data and publication with XePhIR will be done after the acceptation of the original publication. This will enhance the reach of the original research article, enhance visibility and do not interfere with the publication or copyrights of the original article. With XePhIR, we strive to fulfill these objectives and reason that this resource will enhance reproducibility and showcase the appeal and applicability of the zebrafish xenograft model.
Database URL: https://www.xephir.org
Introduction and purpose
Since the transplantation of human metastatic melanoma cells into zebrafish blastula-stage embryos (Danio rerio) by Lee et al. in 2005 (1), the popularity of the zebrafish to model human cancer has been on a steady rise (Figure 1): scientists all over the world incorporate zebrafish as a model for cancer compound screening and utilize the zebrafish xenograft model to address basic questions in tumor biology and metastasis (2–4).
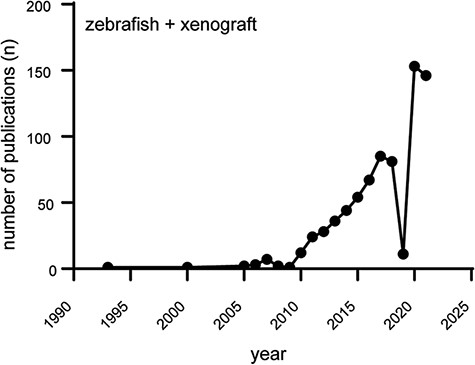
Increasing popularity of the zebrafish xenograft model. Over the course of the last 10 years, the popularity of zebrafish xenograft models has been steadily increasing. Figure based on a https://www.pubmed.gov search for articles containing both the keywords zebrafish and xenograft.
In 2016, we have published the first description of a zebrafish patient-derived xenograft (5). One of the main issues we noted since then was a general lack of conformity among different laboratories working with zebrafish xenograft models (i.e. cell types, growth medium, cancer cell processing prior to engraftment, staining of cells, injection site and time). Recently, several highly translational studies using both stable cell lines and patient-derived xenografts have further supported the overall value of the zebrafish as a cancer model (6–8). One of the main difficulties in evaluating and comparing data obtained with the zebrafish xenograft model as well as interlaboratory adoption is the lack of standardized protocols. Detailed protocols are needed due to the variable nature and needs of each cancer line, which may result in invalid engraftment phenotypes or incorrect data interpretation.
Considering the increasing interest and number of zebrafish-based studies in the cancer field and the ever-growing need for transparent and reproducible science, we have created a platform where zebrafish xenograft phenotypes can be showcased, in an open-access database. This platform will help to exchange experiences inside the zebrafish community as well as increase visibility and appreciation for this useful cancer model by wider audience. The need for a platform of this kind is also evidenced by the recent initiative by Targen et al. 2020, who created a zebrafish xenograft metadata repository (9). However, zebrafish research is often driven by image-based quantification analyses. Therefore, we believe that a visual data repository is a more fitting representation of the underlying data and furthers the goal of enhancing the reproducibility of zebrafish assays. We want to enable external users to quickly choose the cell lines and zebrafish models for their assay of choice, such as dissemination, orthotopic/ectopic engraftment, angiogenesis, etc. (Figure 2). Alternatively, this platform will facilitate the comparison of phenotypes, cell lines and zebrafish models between labs to enhance the overall reproducibility of zebrafish models around the world. In addition, our approach enables easy outreach to users unfamiliar with the zebrafish xenograft model.
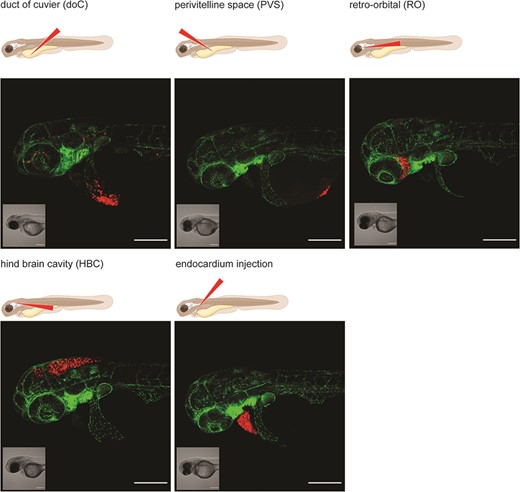
Graphical representation of common injection sites of the zebrafish xenograft model. Injection through the duct of Cuvier, a common route of injection to generate hematogenously disseminated cancer, is also described as an experimental micrometastatic model (10, 11). Perivitelline space injection, originally described as a model for the generation and assessment of angiogenesis, more recently developed as a model for the generation of primary-like tumors (6). Retro-orbital engraftment, used for the generation of orthotopic primary-like tumors derived from eye tumors, allows for the development of distant metastases (12). Hindbrain cavity injection models are used for the generation of orthotopic brain cancer models and for the generation of brain metastasis models (13). Intra-pericardial injection, used for the establishment of primary-like tumors, in an environment closed off from further blood circulation (14, 15). All larvae were injected with ∼200–300, tdTomato expressing cancer cells at 48hpf and imaged at approximately 2 h post-injection using a Zeiss LSM800 laser scanning confocal microscope with an Airyscan detector.
Since most zebrafish assays are image-based, there is an untapped host of data, outlining the zebrafish phenotypes generated during the conventional course of the experiments. We, therefore, propose to use one representative image per timepoint for submission to Xenograft Phenotype Interactive Repository (XePhIR). Citations to the original research will be included to allow and stimulate the citation of the originator of the model to further enhance visibility.
Comprehensive xenograft metadata and phenotype
To enhance the outreach potential of XePhiR, we will include a description of the cancer cells and an explanation of the general experiment (a summary of the goal of the experiment and the outcome <150 words). To facilitate comparison and exchange between laboratories, we will include a comprehensive list of all metadata required to repeat the experiment (cell types, culture conditions, injection space, etc., as specified in Table 1) as well as phenotypes to check if well engrafted. Among the metadata, there are, for example, American Type Cell Culture (ATCC) and/or Expasy (Cellosaurus) identifiers for the engrafted cells, Addgene identifier for reporter constructs used, chemical tracer used (specified, by the supplier and catalog number) and zebrafish line as by Zebrafis information network (ZFIN) identifier. Alternatively, graft or host temperature adaptation can be specified, as temperature variables have been shown to influence engraftment (16).
| Cell line . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Origin | Cellosaurus ID | Medium | Confluence (%) | Culture temp. (°C) | Passage number | Carrier solution | Culture temp. (°C) | ||
| MDA-mb231 | ATCC | CVCL_0062 | DMEM-F12 | 75–90% | 37 | p15 | 2% PVP40 in PBS | 34 | ||
| Dissociation | Injection | |||||||||
| Reagent | Time (min) | Tracer | Construct IDa | Concentration/mL (106) | Timepoint (hpf) | Location | Amount (cells/inj.) | |||
| Trypsin-EDTA | 3–5 min | dsRED | Addgene106171 | 250 | 48 | DoC | 250–350 | |||
| Zebrafish | Phenotype | |||||||||
| Reporter (ZFIN identifier) | synonym | Negative selection | Imaging | |||||||
| ZDB-TGCONSTRCT-070117-94 | Tg(fli1:GFP) | Time (dpi) | Typeb | Timepoints (dpi) | Microscope | |||||
| 1 | CA/YS | 1,4,6 | Leica MZ16FA | |||||||
| Source data | Protocol | |||||||||
| Publication Ref (DOI) | Protocol Ref (DOI) | |||||||||
| doi: 10.3390/ijms17081375 | doi: 10.3791/62356 | |||||||||
| Cell line . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Origin | Cellosaurus ID | Medium | Confluence (%) | Culture temp. (°C) | Passage number | Carrier solution | Culture temp. (°C) | ||
| MDA-mb231 | ATCC | CVCL_0062 | DMEM-F12 | 75–90% | 37 | p15 | 2% PVP40 in PBS | 34 | ||
| Dissociation | Injection | |||||||||
| Reagent | Time (min) | Tracer | Construct IDa | Concentration/mL (106) | Timepoint (hpf) | Location | Amount (cells/inj.) | |||
| Trypsin-EDTA | 3–5 min | dsRED | Addgene106171 | 250 | 48 | DoC | 250–350 | |||
| Zebrafish | Phenotype | |||||||||
| Reporter (ZFIN identifier) | synonym | Negative selection | Imaging | |||||||
| ZDB-TGCONSTRCT-070117-94 | Tg(fli1:GFP) | Time (dpi) | Typeb | Timepoints (dpi) | Microscope | |||||
| 1 | CA/YS | 1,4,6 | Leica MZ16FA | |||||||
| Source data | Protocol | |||||||||
| Publication Ref (DOI) | Protocol Ref (DOI) | |||||||||
| doi: 10.3390/ijms17081375 | doi: 10.3791/62356 | |||||||||
Addgene ID or other stable ID.
CE (cardiac edema), YS (yolk sac injection/leakage), UD (unwanted dissemination) MF (malformation).
| Cell line . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Origin | Cellosaurus ID | Medium | Confluence (%) | Culture temp. (°C) | Passage number | Carrier solution | Culture temp. (°C) | ||
| MDA-mb231 | ATCC | CVCL_0062 | DMEM-F12 | 75–90% | 37 | p15 | 2% PVP40 in PBS | 34 | ||
| Dissociation | Injection | |||||||||
| Reagent | Time (min) | Tracer | Construct IDa | Concentration/mL (106) | Timepoint (hpf) | Location | Amount (cells/inj.) | |||
| Trypsin-EDTA | 3–5 min | dsRED | Addgene106171 | 250 | 48 | DoC | 250–350 | |||
| Zebrafish | Phenotype | |||||||||
| Reporter (ZFIN identifier) | synonym | Negative selection | Imaging | |||||||
| ZDB-TGCONSTRCT-070117-94 | Tg(fli1:GFP) | Time (dpi) | Typeb | Timepoints (dpi) | Microscope | |||||
| 1 | CA/YS | 1,4,6 | Leica MZ16FA | |||||||
| Source data | Protocol | |||||||||
| Publication Ref (DOI) | Protocol Ref (DOI) | |||||||||
| doi: 10.3390/ijms17081375 | doi: 10.3791/62356 | |||||||||
| Cell line . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Origin | Cellosaurus ID | Medium | Confluence (%) | Culture temp. (°C) | Passage number | Carrier solution | Culture temp. (°C) | ||
| MDA-mb231 | ATCC | CVCL_0062 | DMEM-F12 | 75–90% | 37 | p15 | 2% PVP40 in PBS | 34 | ||
| Dissociation | Injection | |||||||||
| Reagent | Time (min) | Tracer | Construct IDa | Concentration/mL (106) | Timepoint (hpf) | Location | Amount (cells/inj.) | |||
| Trypsin-EDTA | 3–5 min | dsRED | Addgene106171 | 250 | 48 | DoC | 250–350 | |||
| Zebrafish | Phenotype | |||||||||
| Reporter (ZFIN identifier) | synonym | Negative selection | Imaging | |||||||
| ZDB-TGCONSTRCT-070117-94 | Tg(fli1:GFP) | Time (dpi) | Typeb | Timepoints (dpi) | Microscope | |||||
| 1 | CA/YS | 1,4,6 | Leica MZ16FA | |||||||
| Source data | Protocol | |||||||||
| Publication Ref (DOI) | Protocol Ref (DOI) | |||||||||
| doi: 10.3390/ijms17081375 | doi: 10.3791/62356 | |||||||||
Addgene ID or other stable ID.
CE (cardiac edema), YS (yolk sac injection/leakage), UD (unwanted dissemination) MF (malformation).
Database architecture
XePhiR has been built in an SQL and front-faced in a website using a custom WordPress installation, which we plan to open source in the future. This front-end database would aggregate and integrate data openly published and citable from their authors using Zenodo or other open-access data repositories. This solution simplifies the long-term maintenance and property rights management of all data shown in XePhIR.
Database contents
Since most zebrafish xenograft experiments are image-based, we have chosen a graphic driven, searchable database. Here, users can upload one or more data sets per timepoint of the performed zebrafish experiment (Fiji/bioformats compatible raw data only) (17). The option to deposit multiple images per timepoint should be used when the established phenotypes clearly segregate, for example, when a graft develops either localized or disseminated tumor growth. The visual data will be supplemented with a comprehensive set of metadata as described previously (representation of the final website user interface in Figure 3). To ensure a high quality of uploaded data, we will curate all final submissions based on proper quality and experimental design. All permissible data will be generated using either fluorescent proteins as cell tracers or will have to be generated using transient chemical tracers (12–19). In the case of chemical tracers, these have to be preferentially followed by anti-human specific immunostaining or in a minimal case with a nuclear dye to show the presence of intact nuclei within the established tumor (18, 19).
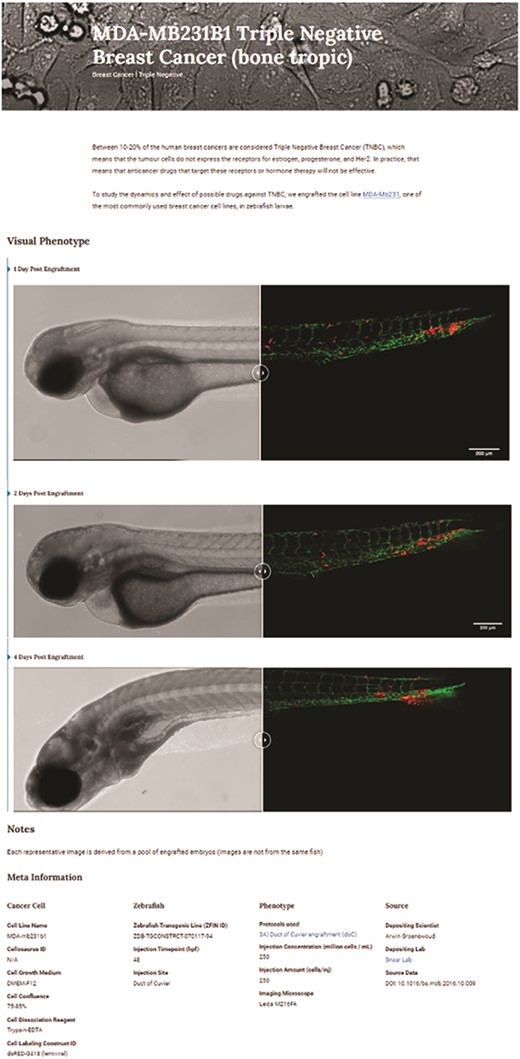
Triple-negative breast cancer sample data as deposited. Image-based analysis data are used to generate a graph for the original publication and are subsequently uploaded to XePHIR (either directly or indirectly). Subsequently, the available metadata is entered during submission (filling in the available data in the submission template). All metadata will be displayed next to the images that have been uploaded, when uploaded via Zenodo a DOI will be provided to allow easy access to the original data set.
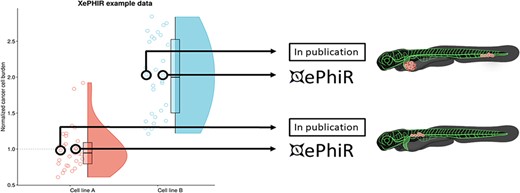
Data are derived from redundant image-based analysis. Image-based analysis data are used to generate a graph and are subsequently used for the publication of the original paper, where one representative sample individual is used in the original manuscript. Another representative individual is submitted to XePHIR either directly or indirectly through deposition of the underlying data set in Zenodo and subsequent publication of the representative image in XePhIR. Using the plotted data used for the generation of a figure in a research article allows for the selection of an adjacent, representative individual in normally distributed data.
Standardized protocols
As to further enhance the ease of interpretation of protocols and to support the worldwide repeatability of zebrafish xenograft experimentation, we have provided standardized engraftment protocols. These protocols are dynamic and can be adapted through drop-down menus to encompass all conventional injection sites (duct of Cuvier, perivitelline space, retro-orbital and hindbrain cavity), carrier solutions, cell densities, cell number and time of injection (in hours post-fertilization, hpf). Furthermore, we will facilitate the uploading of external protocols with each submission (either direct upload or linking out to the protocol when available in an open-access publication or other freely accessible website) to further enhance reproducibility.
Submission guidelines and intellectual property
Our database is open for submission and mainly functions as a graphical front end for externally stored data sets. Alternatively, single-phenotype images per timepoint can be submitted, although linking to whole data sets is preferred, in line with the enhancement of reproducibility and transparency.
Whole data sets will be deposited preferentially in external, freely available data repositories (such as Zenodo). The phenotype(s) to be displayed on XePhIR will be indicated on the submission form, along with the required metadata. Data from previously published manuscripts can be submitted, by selecting a representative image from published data sets, which have not previously been used in print (center of the distribution when plotted, comparable to the image used in the original scientific publication, Figure 5). The selected data will be uploaded to XePhIR after it has been assessed for quality (either using fluorescent proteins or a lipid tracer in conjunction with immunostaining).
The focus of XePhIR will be placed on the dissemination of already published data, to ensure that there will be no conflicts of interest between publishers and researchers. Next to the dissemination of previously published work, we will also provide a platform for unpublished data. Given that this could lead to a possible conflict of interest during later publication in a journal, this can only be done at the users’ informed request. If needed, uploaded data will be time-gated prior to publication, where the items will be released to the public after the publication of the original manuscript.
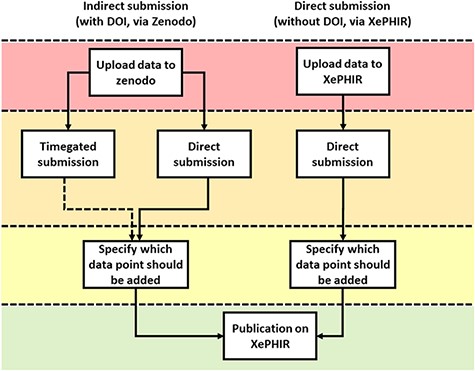
Methods of data submission to XePHIR. Indirect submission through Zenodo, allowing uploading of whole data sets (after acceptation of publication) will allow for enhanced transparency and reusability of data and will provide the user with a DOI-enabling citation of the data set. Uploading to Zenodo will automatically place the data under a CC license. Direct submission to XePHIR will not provide the user with a DOI and will require the user to place the data under a CC license.
Careful selection of the representative data submitted to XePhIR will ensure the nature of the phenotype depicted and will thus enhance global reproducibility and minimize bias when selecting cancer cell lines for future experiments (Figure 4).
As the guidelines for the ethical use of animals vary between different countries, we will ask all depositors to sign a waiver stating that the generation of all data deposited has been generated in accordance with local animal ethical guidelines. We do not condone and take no responsibility for the unethical use of animals.
To submit data to XePhIR, data have to be placed under a selection of creative commons (CC) licenses, namely the choice between the BY non commercial, no derivatives, BY non commercial, share alike and BY share alike licenses (more information at .https://creativecommons.org). All CC licenses chosen disallow respective commercial use. Through CC licensing, we can facilitate the use of the deposited material for future use in, for instance, grant applications. To do so, data can be uploaded either directly to https://www.xephir.org or via Zenodo (20) as a whole data set, thus automatically placing it under CC license and generating a digital object identifier (DOI) allowing the referencing of the data (Figure 5).
Conclusion
With XePhIR, we represent for the first time a visual, user-driven zebrafish xenograft database, which provides one-page summaries with the most important information of a cancer study utilizing zebrafish (Figure 5), collating all required data to reproduce the experiments, referring to the original paper. Through XePhIR, we will enhance the visibility of individual research groups and the zebrafish xenograft models they generate and use. Using a two-pronged data submission approach, we will be able to generate data for a future comparative analysis between groups, models or cell lines, further enhancing the reproducibility of zebrafish xenograft models worldwide. Data submission through Zenodo would further enhance the data reuse capacity and allows for the referencing of individual (un)published data sets using the Zenodo-generated DOI.
Funding
European Union’s Horizon 2020 research and innovation program (667787; UM Cure 2020 project, www.umcure2020.org) to A.G.; Wilhelm Sander-Stiftung (2019.143.1) to F.B.E.
Conflict of interest
None declared.
References
Author notes
contributed equally to this work.



