-
PDF
- Split View
-
Views
-
Cite
Cite
Shijun Zhang, Hengyi Wu, Lei Wang, Gongbo Zhang, Luis M Rocha, Hagit Shatkay, Lang Li, Translational drug–interaction corpus, Database, Volume 2022, 2022, baac031, https://doi.org/10.1093/database/baac031
Close - Share Icon Share
Abstract
The discovery of drug–drug interactions (DDIs) that have a translational impact among in vitro pharmacokinetics (PK), in vivo PK and clinical outcomes depends largely on the quality of the annotated corpus available for text mining. We have developed a new DDI corpus based on an annotation scheme that builds upon and extends previous ones, where an abstract is fragmented and each fragment is then annotated along eight dimensions, namely, focus, polarity, certainty, evidence, directionality, study type, interaction type and mechanism. The guideline for defining these dimensions has undergone refinement during the annotation process. Our DDI corpus comprises 900 positive DDI abstracts and 750 that are not directly relevant to DDI. The abstracts in corpus are separated into eight categories of DDI or non-DDI evidence: DDI with pharmacokinetic (PK) mechanism, in vivo DDI PK, DDI clinical, drug–nutrition interaction, single drug, not drug related, in vitro pharmacodynamic (PD) and case report. Seven annotators, three annotators with drug–interaction research experience and four annotators with less drug–interaction research experience independently annotated the DDI corpus, where two researchers independently annotated each abstract. After two rounds of annotations with additional training in between, agreement improved from (0.79, 0.96, 0.86, 0.70, 0.91, 0.65, 0.78, 0.90) to (0.93, 0.99, 0.96, 0.94, 0.95, 0.93, 0.96, 0.97) for focus, certainty, evidence, study type, interaction type, mechanisms, polarity and direction, respectively. The novice-level annotators improved from 0.83 to 0.96, while the expert-level annotators stayed in high performance with some improvement, from 0.90 to 0.96. In summary, we achieved 96% agreement among each pair of annotators with regard to the eight dimensions. The annotated corpus is now available to the community for inclusion in their text-mining pipelines.
Database URLhttps://github.com/zha204/DDI-Corpus-Database/tree/master/DDI%20corpus
Introduction
Research into drug–drug interactions (DDIs) has been rapidly accelerated in recent years (1). DDIs impact patient safety (2) and healthcare costs (3) and are a major cause of adverse drug reactions. As a result, healthcare information systems have implemented clinical alerts that allow the early detection of DDIs to prevent associated negative outcomes (4). The efficacy of these alerts relies heavily on good curation of knowledge from the literature that provides reliable and comprehensive information about drugs and their interactions (5) as well as their clinical impact and underlying pharmacological mechanisms (6). Such curation depends on effective text-mining tools. Text mining can help identify novel DDI signals from the literature (7, 8), and as such DDI text mining has become an important research area in both pharmacology (7) and informatics (9–12). The development of text-mining methods often requires well-annotated corpus, where corpus construction is guided by documentation, balanced text class composition, recoverability and data on annotator agreement (13). In this paper, we will address how DDI shall be curated and annotated in a corpus. This corpus will be highly valuable for the future text-mining analysis, but the text-mining analysis is not in the scope of this paper.
Various techniques have been considered to represent the text class composition in the context of published science. One scheme, zone partition (14–16), divides the full text of biological articles into 10 zones, background, problem-setting (i.e. the goal of the paper), method, results, insight (i.e. interpretation of the observed data), implication (i.e. implication of the author’s work, such as the applications, limitations and future work), other (i.e. other kind of information of the author’s own work), difference (i.e. statements describing contrasting relations between zones), connection (i.e. statements describing consistent relation between zones) and outline.
A different scheme, which forms the basis to ours and is adopted here (17), emphasized the scientific contents within a fragment of text, either a sentence or a sentence fragment, and examined dimensions including focus = scientific vs. general, polarity—either positive or negative, certainty—the degree of confidence about the validity of the assertion, evidence—whether there is evidential support to the statement, and direction—an increase or a decrease in the level of a phenomenon or an activity.
Three corpora have been specifically developed, pertaining to DDI evidence. In 2011 and 2013, the DDI Extraction Challenge Tasks (DDI-ECT) (18–20) provided annotation of pharmacological substances and DDI relationships from biomedical texts. In the 2011 DDI-ECT, the annotated DDI corpus consists of 579 text passages selected from the Interactions field of DrugBank; in the 2013 DDI-ECT, the corpus was created using 792 text passages selected from the Interactions field of DrugBank and 233 PubMed abstracts. DDI-ECT provided drug annotations by labeling generic name as drug, brand name as brand, group name as group and substrate name as drug_n, the latter representing a substrate of the drug not approved for human use. In addition, DDI-ECT annotated four drug–interaction relationships—the pharmacokinetic (PK) mechanism, mechanism; the pharmacodynamic (PD) effect of drug interaction, effect; advice regarding drug interaction, advice; and the absence of specific information regarding DDI, int, which stands for interaction. The PK DDI package insert corpus (PK-DDI-PI) (21) provided a second body of information regarding evidence for drug interactions and was built utilizing Food and Drug Administration (FDA)-approved drug labels, focusing exclusively on annotating DDI relationships. Initially, it differentiated PK DDIs from PD DDIs. In the case of a PK DDI pair, one drug, referred to as the precipitant, inhibits/induces the pharmacokinetics of another drug, referred to as the object, where the corpus defined the precipitant/object roles in the pair and further labeled each drug in the pair as either an active ingredient, product or metabolite, as well as identified the positive/negative and quantitative/qualitative modalities for the pair.
Our group undertook the development of a third annotation scheme to create a PK corpus that focuses more on the pharmacokinetics of DDIs (22). It first comprised four classes of PK abstracts, including in vivo PK, in vivo pharmacogenetic (23), in vivo DDI and in vitro DDI studies. The annotation scheme had a 3-layer hierarchical structure, in which the first layer annotated entities (drug, dose, enzyme, PK parameter, unit, sample size, mechanism, adjective [adj] word and action word); the second layer annotated sentences concerning DDI (clear DDI, vague DDI or no DDI) and the third layer annotated DDI relationships between two drugs or between a drug and an enzyme with respect to in vivo and in vitro evidence in a sentence. An in vivo DDI pair was labeled DDI, ambiguous DDI (ADDI) or non-DDI, and an in vitro DDI pair was characterized as DDI, drug–enzyme interaction (DEI), ADDI, ambiguous DEI (ADEI), non-DDI or non-DEI. Notably, the label no DDI in the second layer indicates no DDI evidence in a sentence, while the label non-DDI in the third layer indicates that a pair of drugs do not have interaction. Table 1 summarizes the main properties all three DDI corpus and their annotation schemes.
| Corpus . | Data sources . | Annotation schemes . |
|---|---|---|
| DDI-ECT | DrugBank sentences | Entities: drug, brand, group, and drug_n |
| Medline abstracts | DDI relationships: mechanism, effect, advice and inta | |
| PK-DDI-PI | FDA drug labels | DDI relationships: pharmacodynamics/pharmacokinetics, type, role, positive/negative, qualitative/quantitative |
| PK corpus | Medline abstracts | Entities: drug, dose, enzyme, PK parameter, unit, sample size, mechanism, adjective word, action word |
| DDI sentences: clear DDI, vague DDI, no DDIb | ||
| DDI relationships: in vivo DDI (DDI, ambiguous DDI (ADDI), non-DDI) and in vitro DDI (DDI, DEI, ADDI, ADEI, non-DDI, non-DEI) |
| Corpus . | Data sources . | Annotation schemes . |
|---|---|---|
| DDI-ECT | DrugBank sentences | Entities: drug, brand, group, and drug_n |
| Medline abstracts | DDI relationships: mechanism, effect, advice and inta | |
| PK-DDI-PI | FDA drug labels | DDI relationships: pharmacodynamics/pharmacokinetics, type, role, positive/negative, qualitative/quantitative |
| PK corpus | Medline abstracts | Entities: drug, dose, enzyme, PK parameter, unit, sample size, mechanism, adjective word, action word |
| DDI sentences: clear DDI, vague DDI, no DDIb | ||
| DDI relationships: in vivo DDI (DDI, ambiguous DDI (ADDI), non-DDI) and in vitro DDI (DDI, DEI, ADDI, ADEI, non-DDI, non-DEI) |
Note: inta means interaction; no DDIb means no drug interaction evidence in a sentence.
| Corpus . | Data sources . | Annotation schemes . |
|---|---|---|
| DDI-ECT | DrugBank sentences | Entities: drug, brand, group, and drug_n |
| Medline abstracts | DDI relationships: mechanism, effect, advice and inta | |
| PK-DDI-PI | FDA drug labels | DDI relationships: pharmacodynamics/pharmacokinetics, type, role, positive/negative, qualitative/quantitative |
| PK corpus | Medline abstracts | Entities: drug, dose, enzyme, PK parameter, unit, sample size, mechanism, adjective word, action word |
| DDI sentences: clear DDI, vague DDI, no DDIb | ||
| DDI relationships: in vivo DDI (DDI, ambiguous DDI (ADDI), non-DDI) and in vitro DDI (DDI, DEI, ADDI, ADEI, non-DDI, non-DEI) |
| Corpus . | Data sources . | Annotation schemes . |
|---|---|---|
| DDI-ECT | DrugBank sentences | Entities: drug, brand, group, and drug_n |
| Medline abstracts | DDI relationships: mechanism, effect, advice and inta | |
| PK-DDI-PI | FDA drug labels | DDI relationships: pharmacodynamics/pharmacokinetics, type, role, positive/negative, qualitative/quantitative |
| PK corpus | Medline abstracts | Entities: drug, dose, enzyme, PK parameter, unit, sample size, mechanism, adjective word, action word |
| DDI sentences: clear DDI, vague DDI, no DDIb | ||
| DDI relationships: in vivo DDI (DDI, ambiguous DDI (ADDI), non-DDI) and in vitro DDI (DDI, DEI, ADDI, ADEI, non-DDI, non-DEI) |
Note: inta means interaction; no DDIb means no drug interaction evidence in a sentence.
Comparison of the general guidelines for corpus annotation with the annotation schemes used for existing DDI corpus reveals several obvious gaps. The aforementioned segmentation of most current publications into background, materials and methods, results and conclusions sections has rendered the zone partition scheme redundant (14–16); current DDI corpus does not utilize the five dimensions of focus, polarity, certainty, strength and direction (17), although they offer a much in-depth annotation of drug pharmacology.
Here we report our efforts combining the advantages of multiple annotation guidelines to construct a corpus that allows sufficient representation and generalizability of scientific knowledge, while maintaining the dimensions of focus, polarity, certainty, evidence and directionality, and includes DDI study type, mechanism and interaction type to integrate sufficient science, i.e. pharmacology, within the annotation scheme.
Materials and methods
Screening of abstracts for corpus inclusion
Our drug–interaction corpus comprises abstracts from PubMed that we screened via a keyword query [‘drug interaction’ AND (Type of Study)]. We established abstract selection criteria (Table 2) to classify the abstracts included in our corpus into eight subcategories, three for DDI abstracts, including PK in vitro, PK in vivo and clinical, and five for non-DDI abstracts, including drug–nutrition interaction, single drug, not drug related, in vitro PD and case report. Abstract classification was performed by four annotators, two experienced ones and two novices. Two experienced annotators included one with a Master’s degree in biology and 3-year experience in drug–interaction corpus development. The second had a Ph.D. in bioinformatics with pharmacology training background, and his Ph.D. thesis topic focused on PK and drug–interaction corpus development and text mining. The other two novice annotators had either biology or pharmacology background. When they participated in this annotation project, they just started their training programs in bioinformatics. All the abstracts were classified by these four annotators independently, and each abstract was labeled by two annotators. The disagreed annotations were further validated by two experienced annotators jointly for the final decision (see Figure 1).
| Study type . | Description . |
|---|---|
| PK in vitro drug interaction | Substrate depletion or metabolite formation studies between substrate drugs and probe inhibitor drugs for metabolism enzymes or transporters; and inhibition/induction studies between inhibitor/inducer drugs and probe substrate drugs for metabolism enzymes or transporters |
| PK in vivo drug interaction | Clinical pharmacokinetics studies that compare a substrate drug’s exposure alone to the substrate drug exposure co-committed with inhibitor or inducer drugs |
| Clinical drug interaction | Phase I/II/III clinical trials with reported drug combination and/or single drug toxicity data. Pharmaco-epidemiology studies with reported toxicities from drug combinations |
| Drug–nutrition interaction | The interaction studies between drugs and natural products, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| Single drug | Single drug studies, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| No drug related | Clinical or preclinical studies, but not drug related |
| Case report | Single drug or drug combination induced Adverse drug effect cases |
| PD in vitro | Cell culture studies on pharmacodynamics, but not on pharmacokinetics |
| Study type . | Description . |
|---|---|
| PK in vitro drug interaction | Substrate depletion or metabolite formation studies between substrate drugs and probe inhibitor drugs for metabolism enzymes or transporters; and inhibition/induction studies between inhibitor/inducer drugs and probe substrate drugs for metabolism enzymes or transporters |
| PK in vivo drug interaction | Clinical pharmacokinetics studies that compare a substrate drug’s exposure alone to the substrate drug exposure co-committed with inhibitor or inducer drugs |
| Clinical drug interaction | Phase I/II/III clinical trials with reported drug combination and/or single drug toxicity data. Pharmaco-epidemiology studies with reported toxicities from drug combinations |
| Drug–nutrition interaction | The interaction studies between drugs and natural products, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| Single drug | Single drug studies, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| No drug related | Clinical or preclinical studies, but not drug related |
| Case report | Single drug or drug combination induced Adverse drug effect cases |
| PD in vitro | Cell culture studies on pharmacodynamics, but not on pharmacokinetics |
| Study type . | Description . |
|---|---|
| PK in vitro drug interaction | Substrate depletion or metabolite formation studies between substrate drugs and probe inhibitor drugs for metabolism enzymes or transporters; and inhibition/induction studies between inhibitor/inducer drugs and probe substrate drugs for metabolism enzymes or transporters |
| PK in vivo drug interaction | Clinical pharmacokinetics studies that compare a substrate drug’s exposure alone to the substrate drug exposure co-committed with inhibitor or inducer drugs |
| Clinical drug interaction | Phase I/II/III clinical trials with reported drug combination and/or single drug toxicity data. Pharmaco-epidemiology studies with reported toxicities from drug combinations |
| Drug–nutrition interaction | The interaction studies between drugs and natural products, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| Single drug | Single drug studies, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| No drug related | Clinical or preclinical studies, but not drug related |
| Case report | Single drug or drug combination induced Adverse drug effect cases |
| PD in vitro | Cell culture studies on pharmacodynamics, but not on pharmacokinetics |
| Study type . | Description . |
|---|---|
| PK in vitro drug interaction | Substrate depletion or metabolite formation studies between substrate drugs and probe inhibitor drugs for metabolism enzymes or transporters; and inhibition/induction studies between inhibitor/inducer drugs and probe substrate drugs for metabolism enzymes or transporters |
| PK in vivo drug interaction | Clinical pharmacokinetics studies that compare a substrate drug’s exposure alone to the substrate drug exposure co-committed with inhibitor or inducer drugs |
| Clinical drug interaction | Phase I/II/III clinical trials with reported drug combination and/or single drug toxicity data. Pharmaco-epidemiology studies with reported toxicities from drug combinations |
| Drug–nutrition interaction | The interaction studies between drugs and natural products, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| Single drug | Single drug studies, including in vitro PK experiments, clinical PK studies and clinical/epidemiology studies on efficacy or toxicity |
| No drug related | Clinical or preclinical studies, but not drug related |
| Case report | Single drug or drug combination induced Adverse drug effect cases |
| PD in vitro | Cell culture studies on pharmacodynamics, but not on pharmacokinetics |
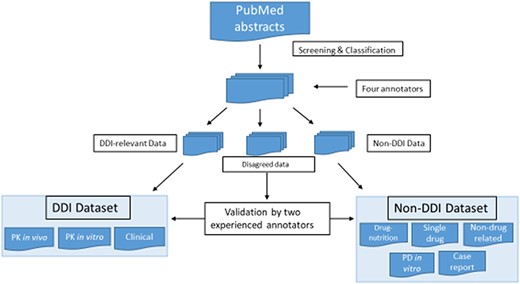
Screening and classification of PubMed abstracts for corpus development. PubMed abstracts were initially screened by four annotators into DDI-relevant, non-DDI and disagreed data. The four annotators included two experienced ones and two novice ones. The two experienced annotators had drug–interaction research experience, and two novice annotators were new bioinformatics master novices with biology and pharmacology background. All the abstracts were classified by these four annotators independently, and each abstract was labeled by two annotators. The disagreed data were further validated by two experienced annotators.
Building the drug–interaction corpus
Annotator characteristics
Our expert group comprised three ‘experienced annotators’. The first one has a Master’s degree in biology with 3-year experience in drug–interaction corpus development and text mining. The second has a Ph.D. in bioinformatics with pharmacology training background; the Ph.D. thesis topic focused on PK and drug–interaction corpus development. The third is a second-year Master’s novice in bioinformatics with a Master’s degree in biology and 1.5-year drug–interaction corpus annotation experience in the lab. Four additional annotators were ‘novice annotators’ with either biology or pharmacology or informatics background. When they participated into this annotation project, they just started their training programs in the Indiana University. One is a new research follow in clinical pharmacology, who had pharmacology training background, but no informatics expertise. The other three are new master novices in bioinformatics. The three experienced annotators were involved in developing the annotation guidelines. These three experienced annotators trained the novice annotators. Note that four of the seven corpus annotators were also the ones who classified the abstracts in the previous step (Figure 1).
Annotation process
After the identification of positive and negative DDI abstracts in the screening stage, the novice annotators read the annotation guidelines and underwent training and practiced annotation at the sentence level, applying the guidelines to five abstracts as test examples. Following that training stage, in a first round of annotations, abstracts in the corpus were randomly assigned for independent annotation by each of two annotators. The novice annotators then received additional training that involved discussion of any inconsistencies or conflicts in their understanding and application of the guidelines. In a second round of annotation, the annotators reviewed and revised their first annotations as necessary. In cases of disagreement after the second round, one of the three experienced annotators provided the final annotation. Figure 2 illustrates the sentence-level annotation process.
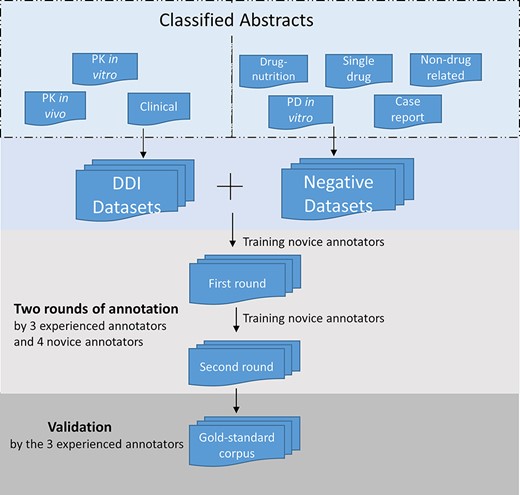
Annotation process for the corpus development. The DDI corpus was constructed by three experienced annotators and four novice annotators. Novice annotators received two rounds of training of sentence-level annotation. After the initial training, each abstract in the corpus was assigned randomly to two annotators for the first-round annotation. Then the novice annotators underwent additional training. In the second-round annotation, the annotators reviewed and revised their annotation. The annotations were validated and finalized by the experienced annotators.
Annotation guidelines
Building on the annotation guidelines previously developed by Dr Hagit Shatkay and her colleagues [17], the annotation guidelines aim to identify drug–interaction entities and evidence of such interaction within the pharmacology-related literature. The guidelines delineate the rules and conventions for conducting the annotation task and provide case examples. The unit of annotation is a fragment within a sentence. A sentence is fragmented whenever there is a change in the annotation value along any of the eight dimensions, namely, focus, polarity, certainty, evidence, directionality, study type, interaction type or DDI mechanisms.
Focus Each fragment may convey one or more of the following categories:
Scientific content, findings or discovery. We define this type of information as science and annotate it with the tag S. The tag S is assigned to most sentences that describe prospective or future study.
Generic information. This category refers to a general statement of knowledge and science that is outside the scope of the paper, the structure of the paper itself or the statement of the research world. Such statement is usually not based on scientific experimentation and can reflect an opinion or an observation that would probably be considered true or valid if made by a layperson. We denote generic information using the tag G.
Methodology. This designation refers to methods used in a biological or pharmacological experiment or employed in a clinical study and assigned the tag M.
The focus of a fragment is contingent on context. What may be regarded as a scientific finding in one context may be considered methodology in another. We only annotated methodology when the fragment contained an indication that methodology is discussed. In some cases, a fragment discussing methodology may also discuss science. In such cases, both tags, M and S, are assigned. Other cases may require other tag combinations, such as GS, GM or GMS.
Polarity A fragment can be stated either positively (P) or negatively (N). For instance, the phrase ‘No influence’ in the sentence ‘No influence of cimetidine was observed on the kinetics of single doses of femoxetine.’ indicates a negative polarity (N). As another example, the sentence ‘After multiple doses the plasma concentration of femoxetine was significantly increased.’ shows a positive polarity (P). If the polarity is not clearly stated, for example, ‘It is still unknown whether…’, the fragment is tagged P.
Certainty Each fragment conveys a degree of certainty about the validity of the assertion it makes, which the annotation grades on a scale from 0 to 3. The lowest degree (0) represents complete uncertainty; that is, the fragment explicitly states that there is an uncertainty or lack of knowledge about a particular phenomenon (e.g. ‘it is unknown…’ or ‘it is unclear whether…’). The highest degree (3) represents complete certainty, reflecting an accepted, known and/or proven fact. The intermediate degree (1) reflects low certainty and (2) reflects expressions with high likelihood that are still short of complete certainty.
Evidence This dimension denotes the presence or absence of evidence to support the assertion expressed in the fragment, regardless of the fragment’s focus or certainty. This category is denoted by a tag starting with the letter E followed by one or more digits from 0 to 3 to indicate the evidence type or the letter N to indicate the provision of numerical evidence within the fragment:
E0 indicates that there is either no evidence in the fragment or an explicit statement in the text indicates the absence of evidence (‘ICG-001 binds specifically to CBP…’).
E1 indicates a claim of evidence without explicit information to verify the claim. The fragment does not demonstrate evidence, and there is no explicit reference to evidence. The evidence is merely asserted to exist in some form, possibly in the preceding text or in prior experiments, but its location is not stated. Note that in this case the indirect implication of evidence may not be provided in the fragment, but the use of terms referring to a previous fragment may imply evidence (‘Previous studies suggest that ICG-001 binds specifically to CBP…’).
E2 signifies the absence of evidence within the sentence/fragment, but the presence of explicit reference to other papers (citations) to support the assertion of evidence (‘Previous studies suggest that ICG-001 binds specifically to CBP…[25]’).
E3 represents the presence of evidence within the fragment in one of the following forms:
reference to experiments previously reported within the body of the paper by a direct description of the findings as experimental results (‘Our data demonstrate…’);
use of a verb (typically in the past tense) within the statement that indicates an observation or experimental finding that is described within the paper. For example, ‘We found that…’, ‘We see that…’ and ‘The level of …increased over time…’; and
reference to an experimental figure or table of data given within the paper.
EN denotes the provision of evidence as the numerical results of the experiment described within the paper, such values as PK/PD parameters, sample size, drug doses and treatment time (‘Omeprazole had no apparent effect on the mean (S)-warfarin plasma concentration (379 ng/ml with, versus 387 ng/ml without, omeprazole), …’).
Direction/Trend A plus sign (+) indicates a qualitatively increased level in a specific phenomenon, finding or activity, whereas a minus sign (–) indicates a reduced level. This tag is introduced to separate the notion of positive/negative results and assertions from the level of the observed phenomenon itself. For example, the fragment ‘Nitrendipine 20 mg daily led to a significant increase in plasma digoxin levels and’ is annotated with the tag ‘+’ as it discusses an increase in plasman digoxin levels, while the sentence ‘AUC (0,24 h) of digoxin, however, was slightly reduced after 1 week of treatment with bosentan.’ receives the tag ‘-’ as it discusses a reduction in digoxin.
If both reduction and increase of the same phenomenon are presented as possible, the fragment is tagged by ±. For example, in the sentence ‘Pharmacokinetic drug interactions may result in a decrease or increase in the oral bioavailability of some drugs.’, the change of the oral bioavailability can be either an increase or a decrease, and the fragment is annotated by ±.
Study type Each fragment may contain information such as experimental method or endpoint that can indicate a certain study type, which is indicated by a tag starting with the letter V. A subsequent letter indicates the type of study or its absence. We define five study types that can be associated with a fragment: in vivo (VV), in vitro (VT), clinical (VC), do not know (V0) and not applicable (VN).
The study type of the studies that have no drugs in them is annotated as VN.
VV studies are those in which the pharmacokinetics entities of drugs are tested on humans through clinical studies.
VT studies are those in which the pharmacokinetics (PK) and pharmacodynamics (PD) entities of drugs are tested on cell or human liver microsome models.
VC studies are those in which the pharmacodynamics entities of drugs are tested in clinical studies, including randomized clinical trials, prospective or retrospective cohort studies and case/control studies. The endpoints of clinical studies are the identification of disease symptoms and adverse drug effects, and these endpoints are usually measured according to their likelihood and characterized as odds or hazard ratios or other risk statistics.
When a fragment discusses more than one type of study, the multiple studies are annotated with the tags for each type separated by ‘|’. Thus, the discussion of a VV study and a VC study would be tagged VV|VC.
The tag V0 is assigned when the study type is stated ambiguously and there is no explicit evidence to indicate whether the fragment is discussing a VV study or a VS study.
Interaction type indicates the relationship between drugs and drugs/enzymes occurring in the fragment. The types of interactions include single drug (DR), drug–enzyme interaction (DE), DDI (24) and no drug discussed (D0). The tag DR is assigned to indicate that the description of a drug–drug pair or a drug–enzyme pair in the fragment does not specify an explicit interaction between them. If a fragment indicates both DDIs and drug–enzyme interactions, it will be tagged DD|DE.
Mechanism represents the mechanism of DDI or drug–enzyme interaction. Our annotation uses the labels inhibition (MI), induction (MD), metabolism (MM), transport (5), synergism (25), antagonism (MN), additive (MA) and not applicable (M0) to differentiate mechanism types. When a statement indicates more than one mechanism of drug–drug or drug–enzyme interaction, we allow a combination of tags, e.g. MI|MM.
Thus, a typical fragment annotation consists of a tag of the form:
**[<Integer>][G|M|S]+[P|N][0–3][E[0|N|1 |2|3]] [-|+] ? [() [VV|VT|VC|V0]* [DR|DE|DD|D0] *[MI|MD|MM|MT|MS|MN|MA|M0]* [ ]]
<Integer> is the ordinal number of the fragment within its sentence, starting at 1.
For instance:
Thus, in this case, the chemistry of the product is similar to that of the signal molecules, **1GP3E1(VND0M0) but there is no complementary relationship to the signal sequences. **2GN3E0(VND0M0)
Treatment of human hepatocytes for 72 h with 2–200 microM thiabendazole produced concentration- dependent increases in CYP1A2, CYP2B6 and CYP3A4 mRNA levels, whereas treatment with butylated hydroxytoluene increased CYP2B6 and CYP3A4 mRNA levels. **1SMP3EN+(VTDEM0)
The effect of two different doses of nitrendipine on plasma digoxin levels, urinary recovery and systolic time intervals was investigated in eight healthy volunteers. **1SP0EN([VV|VC]DDM0)
Effect of saquinavir–ritonavir on cytochrome P450 3A4 activity in healthy volunteers using midazolam as a probe. **1SP3E1(V0[DD|DE]M0)
Quality control analysis
IAA fragmentation is also calculated for eight individual subcategories of abstracts defined in Table 2, in which IAA is calculated among all the sentences in an abstract subcategory in the corpus. The annotator groups include experienced annotators, novice annotators, between experienced and novice annotators and all annotators.
Results
Drug–interaction corpus
Our DDI corpus consists of 1650 abstracts, of which 900 discuss DDIs (referred to as DDI) and 750 do not discuss DDI (referred to as non-DDI). The positive set included 300 abstracts each of three types of DDI studies (see descriptions in Table 2): in vivo DDI PK studies, in vitro DDI PK studies and clinical DDI studies. The negative set comprised five types of studies: in vitro pharmacodynamics (PD) studies (n = 100); drug–nutrition interaction studies (n = 200); single-drug studies (n = 200); clinical case reports (n = 50) and nondrug studies (n = 200). We did not include animal studies. Table 3 summarizes our corpus characteristics and the number of fragments within each subcategory of abstracts; Table 4 presents the annotation distribution along the eight dimensions across the eight study categories.
| Corpus . | Abstracts discussed DDI (DDI) . | Abstracts not discussed DDI (non-DDI) . | ||||||
|---|---|---|---|---|---|---|---|---|
| Study categories | In vivo DDI PK | In vitro DDI PK | DDI Clinical | In vitro PD | Drug–nutrition | Single drug | Case reports | Nondrug studies |
| Abstracts | 300 | 300 | 300 | 100 | 200 | 200 | 50 | 200 |
| # Fragments | 4010 | 3176 | 4041 | 760 | 2871 | 2540 | 564 | 2538 |
| Corpus . | Abstracts discussed DDI (DDI) . | Abstracts not discussed DDI (non-DDI) . | ||||||
|---|---|---|---|---|---|---|---|---|
| Study categories | In vivo DDI PK | In vitro DDI PK | DDI Clinical | In vitro PD | Drug–nutrition | Single drug | Case reports | Nondrug studies |
| Abstracts | 300 | 300 | 300 | 100 | 200 | 200 | 50 | 200 |
| # Fragments | 4010 | 3176 | 4041 | 760 | 2871 | 2540 | 564 | 2538 |
| Corpus . | Abstracts discussed DDI (DDI) . | Abstracts not discussed DDI (non-DDI) . | ||||||
|---|---|---|---|---|---|---|---|---|
| Study categories | In vivo DDI PK | In vitro DDI PK | DDI Clinical | In vitro PD | Drug–nutrition | Single drug | Case reports | Nondrug studies |
| Abstracts | 300 | 300 | 300 | 100 | 200 | 200 | 50 | 200 |
| # Fragments | 4010 | 3176 | 4041 | 760 | 2871 | 2540 | 564 | 2538 |
| Corpus . | Abstracts discussed DDI (DDI) . | Abstracts not discussed DDI (non-DDI) . | ||||||
|---|---|---|---|---|---|---|---|---|
| Study categories | In vivo DDI PK | In vitro DDI PK | DDI Clinical | In vitro PD | Drug–nutrition | Single drug | Case reports | Nondrug studies |
| Abstracts | 300 | 300 | 300 | 100 | 200 | 200 | 50 | 200 |
| # Fragments | 4010 | 3176 | 4041 | 760 | 2871 | 2540 | 564 | 2538 |
| Corpus . | DDI . | Non-DDI . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study categories . | In vivo DDI PK . | In vitro DDI PK . | DDI Clinical . | In vitro PD . | Drug–Nutrition . | Single Drug . | Case Reports . | Nondrug Studies . | |
| Focus | G | 226 | 144 | 272 | 110 | 263 | 253 | 68 | 306 |
| M | 976 | 218 | 944 | 80 | 664 | 374 | 46 | 382 | |
| S | 2740 | 2670 | 2725 | 531 | 1861 | 1785 | 440 | 1727 | |
| S|M | 65 | 141 | 90 | 38 | 90 | 127 | 10 | 120 | |
| S|G | 486 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | |
| G|M | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 3 | |
| Polarity | P | 3325 | 2868 | 3510 | 727 | 2481 | 2217 | 502 | 2319 |
| N | 685 | 308 | 532 | 33 | 400 | 323 | 62 | 219 | |
| Certainty | 0 | 340 | 315 | 373 | 71 | 244 | 220 | 9 | 198 |
| 1 | 20 | 31 | 17 | 1 | 9 | 11 | 3 | 5 | |
| 2 | 319 | 319 | 288 | 76 | 187 | 219 | 85 | 280 | |
| 3 | 3331 | 2521 | 3364 | 612 | 2441 | 2090 | 467 | 2055 | |
| Evidence | E0 | 319 | 172 | 209 | 39 | 103 | 140 | 61 | 232 |
| E1 | 214 | 197 | 276 | 123 | 113 | 127 | 55 | 202 | |
| E2 | 1 | 2 | 1 | 2 | 0 | 2 | 0 | 3 | |
| E3 | 2017 | 2028 | 2195 | 553 | 1669 | 1551 | 350 | 1567 | |
| EN | 1371 | 777 | 1361 | 43 | 996 | 720 | 98 | 534 | |
| Direction | + | 488 | 194 | 607 | 121 | 379 | 258 | 41 | 278 |
| − | 389 | 185 | 359 | 75 | 313 | 248 | 59 | 163 | |
| [+|−] | 2 | 2 | 14 | 0 | 1 | 4 | 0 | 1 | |
| Study Type | VV | 1818 | 93 | 159 | 10 | 996 | 359 | 54 | 14 |
| VT | 49 | 1499 | 22 | 396 | 163 | 732 | 0 | 133 | |
| VC | 300 | 46 | 2620 | 17 | 440 | 534 | 279 | 584 | |
| V0 | 1673 | 1417 | 1097 | 233 | 1125 | 852 | 196 | 585 | |
| VN | 92 | 67 | 77 | 97 | 54 | 27 | 29 | 1203 | |
| VV|VC | 72 | 5 | 64 | 0 | 95 | 21 | 5 | 1 | |
| VV|VT | 5 | 46 | 2 | 6 | 6 | 15 | 1 | 16 | |
| VT|VC | 1 | 3 | 0 | 1 | 2 | 0 | 0 | 2 | |
| Interaction | D0 | 841 | 548 | 1269 | 291 | 1048 | 953 | 174 | 2327 |
| Type | DR | 1131 | 857 | 1321 | 279 | 1584 | 1265 | 218 | 201 |
| DD | 1871 | 527 | 1398 | 187 | 155 | 25 | 149 | 5 | |
| DE | 144 | 1182 | 40 | 2 | 90 | 275 | 19 | 5 | |
| DD|DE | 23 | 162 | 14 | 1 | 4 | 13 | 4 | 0 | |
| Mechanism | M0 | 3692 | 1177 | 3851 | 602 | 2565 | 1963 | 526 | 2334 |
| MI | 164 | 613 | 69 | 47 | 107 | 315 | 7 | 56 | |
| MD | 55 | 57 | 41 | 36 | 34 | 60 | 9 | 60 | |
| MM | 64 | 880 | 31 | 3 | 90 | 155 | 14 | 16 | |
| MT | 9 | 10 | 4 | 14 | 23 | 0 | 1 | 13 | |
| MS | 1 | 2 | 15 | 33 | 3 | 2 | 0 | 38 | |
| MN | 1 | 4 | 4 | 0 | 0 | 3 | 0 | 1 | |
| MA | 2 | 1 | 10 | 1 | 2 | 0 | 0 | 4 | |
| MD|MM | 2 | 12 | 0 | 0 | 6 | 1 | 0 | 1 | |
| MD|MT | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| MD|MI | 12 | 17 | 1 | 10 | 5 | 18 | 2 | 4 | |
| MD|MN | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MS | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| MI|MS | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 1 | |
| MI|MN | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| MI|MT | 4 | 5 | 3 | 1 | 6 | 0 | 2 | 0 | |
| MI|MM | 2 | 391 | 8 | 1 | 28 | 23 | 3 | 0 | |
| MM|MT | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MM|MN | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| MA|MS | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MA|MI | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| MS|MT | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Corpus . | DDI . | Non-DDI . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study categories . | In vivo DDI PK . | In vitro DDI PK . | DDI Clinical . | In vitro PD . | Drug–Nutrition . | Single Drug . | Case Reports . | Nondrug Studies . | |
| Focus | G | 226 | 144 | 272 | 110 | 263 | 253 | 68 | 306 |
| M | 976 | 218 | 944 | 80 | 664 | 374 | 46 | 382 | |
| S | 2740 | 2670 | 2725 | 531 | 1861 | 1785 | 440 | 1727 | |
| S|M | 65 | 141 | 90 | 38 | 90 | 127 | 10 | 120 | |
| S|G | 486 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | |
| G|M | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 3 | |
| Polarity | P | 3325 | 2868 | 3510 | 727 | 2481 | 2217 | 502 | 2319 |
| N | 685 | 308 | 532 | 33 | 400 | 323 | 62 | 219 | |
| Certainty | 0 | 340 | 315 | 373 | 71 | 244 | 220 | 9 | 198 |
| 1 | 20 | 31 | 17 | 1 | 9 | 11 | 3 | 5 | |
| 2 | 319 | 319 | 288 | 76 | 187 | 219 | 85 | 280 | |
| 3 | 3331 | 2521 | 3364 | 612 | 2441 | 2090 | 467 | 2055 | |
| Evidence | E0 | 319 | 172 | 209 | 39 | 103 | 140 | 61 | 232 |
| E1 | 214 | 197 | 276 | 123 | 113 | 127 | 55 | 202 | |
| E2 | 1 | 2 | 1 | 2 | 0 | 2 | 0 | 3 | |
| E3 | 2017 | 2028 | 2195 | 553 | 1669 | 1551 | 350 | 1567 | |
| EN | 1371 | 777 | 1361 | 43 | 996 | 720 | 98 | 534 | |
| Direction | + | 488 | 194 | 607 | 121 | 379 | 258 | 41 | 278 |
| − | 389 | 185 | 359 | 75 | 313 | 248 | 59 | 163 | |
| [+|−] | 2 | 2 | 14 | 0 | 1 | 4 | 0 | 1 | |
| Study Type | VV | 1818 | 93 | 159 | 10 | 996 | 359 | 54 | 14 |
| VT | 49 | 1499 | 22 | 396 | 163 | 732 | 0 | 133 | |
| VC | 300 | 46 | 2620 | 17 | 440 | 534 | 279 | 584 | |
| V0 | 1673 | 1417 | 1097 | 233 | 1125 | 852 | 196 | 585 | |
| VN | 92 | 67 | 77 | 97 | 54 | 27 | 29 | 1203 | |
| VV|VC | 72 | 5 | 64 | 0 | 95 | 21 | 5 | 1 | |
| VV|VT | 5 | 46 | 2 | 6 | 6 | 15 | 1 | 16 | |
| VT|VC | 1 | 3 | 0 | 1 | 2 | 0 | 0 | 2 | |
| Interaction | D0 | 841 | 548 | 1269 | 291 | 1048 | 953 | 174 | 2327 |
| Type | DR | 1131 | 857 | 1321 | 279 | 1584 | 1265 | 218 | 201 |
| DD | 1871 | 527 | 1398 | 187 | 155 | 25 | 149 | 5 | |
| DE | 144 | 1182 | 40 | 2 | 90 | 275 | 19 | 5 | |
| DD|DE | 23 | 162 | 14 | 1 | 4 | 13 | 4 | 0 | |
| Mechanism | M0 | 3692 | 1177 | 3851 | 602 | 2565 | 1963 | 526 | 2334 |
| MI | 164 | 613 | 69 | 47 | 107 | 315 | 7 | 56 | |
| MD | 55 | 57 | 41 | 36 | 34 | 60 | 9 | 60 | |
| MM | 64 | 880 | 31 | 3 | 90 | 155 | 14 | 16 | |
| MT | 9 | 10 | 4 | 14 | 23 | 0 | 1 | 13 | |
| MS | 1 | 2 | 15 | 33 | 3 | 2 | 0 | 38 | |
| MN | 1 | 4 | 4 | 0 | 0 | 3 | 0 | 1 | |
| MA | 2 | 1 | 10 | 1 | 2 | 0 | 0 | 4 | |
| MD|MM | 2 | 12 | 0 | 0 | 6 | 1 | 0 | 1 | |
| MD|MT | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| MD|MI | 12 | 17 | 1 | 10 | 5 | 18 | 2 | 4 | |
| MD|MN | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MS | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| MI|MS | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 1 | |
| MI|MN | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| MI|MT | 4 | 5 | 3 | 1 | 6 | 0 | 2 | 0 | |
| MI|MM | 2 | 391 | 8 | 1 | 28 | 23 | 3 | 0 | |
| MM|MT | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MM|MN | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| MA|MS | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MA|MI | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| MS|MT | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Corpus . | DDI . | Non-DDI . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study categories . | In vivo DDI PK . | In vitro DDI PK . | DDI Clinical . | In vitro PD . | Drug–Nutrition . | Single Drug . | Case Reports . | Nondrug Studies . | |
| Focus | G | 226 | 144 | 272 | 110 | 263 | 253 | 68 | 306 |
| M | 976 | 218 | 944 | 80 | 664 | 374 | 46 | 382 | |
| S | 2740 | 2670 | 2725 | 531 | 1861 | 1785 | 440 | 1727 | |
| S|M | 65 | 141 | 90 | 38 | 90 | 127 | 10 | 120 | |
| S|G | 486 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | |
| G|M | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 3 | |
| Polarity | P | 3325 | 2868 | 3510 | 727 | 2481 | 2217 | 502 | 2319 |
| N | 685 | 308 | 532 | 33 | 400 | 323 | 62 | 219 | |
| Certainty | 0 | 340 | 315 | 373 | 71 | 244 | 220 | 9 | 198 |
| 1 | 20 | 31 | 17 | 1 | 9 | 11 | 3 | 5 | |
| 2 | 319 | 319 | 288 | 76 | 187 | 219 | 85 | 280 | |
| 3 | 3331 | 2521 | 3364 | 612 | 2441 | 2090 | 467 | 2055 | |
| Evidence | E0 | 319 | 172 | 209 | 39 | 103 | 140 | 61 | 232 |
| E1 | 214 | 197 | 276 | 123 | 113 | 127 | 55 | 202 | |
| E2 | 1 | 2 | 1 | 2 | 0 | 2 | 0 | 3 | |
| E3 | 2017 | 2028 | 2195 | 553 | 1669 | 1551 | 350 | 1567 | |
| EN | 1371 | 777 | 1361 | 43 | 996 | 720 | 98 | 534 | |
| Direction | + | 488 | 194 | 607 | 121 | 379 | 258 | 41 | 278 |
| − | 389 | 185 | 359 | 75 | 313 | 248 | 59 | 163 | |
| [+|−] | 2 | 2 | 14 | 0 | 1 | 4 | 0 | 1 | |
| Study Type | VV | 1818 | 93 | 159 | 10 | 996 | 359 | 54 | 14 |
| VT | 49 | 1499 | 22 | 396 | 163 | 732 | 0 | 133 | |
| VC | 300 | 46 | 2620 | 17 | 440 | 534 | 279 | 584 | |
| V0 | 1673 | 1417 | 1097 | 233 | 1125 | 852 | 196 | 585 | |
| VN | 92 | 67 | 77 | 97 | 54 | 27 | 29 | 1203 | |
| VV|VC | 72 | 5 | 64 | 0 | 95 | 21 | 5 | 1 | |
| VV|VT | 5 | 46 | 2 | 6 | 6 | 15 | 1 | 16 | |
| VT|VC | 1 | 3 | 0 | 1 | 2 | 0 | 0 | 2 | |
| Interaction | D0 | 841 | 548 | 1269 | 291 | 1048 | 953 | 174 | 2327 |
| Type | DR | 1131 | 857 | 1321 | 279 | 1584 | 1265 | 218 | 201 |
| DD | 1871 | 527 | 1398 | 187 | 155 | 25 | 149 | 5 | |
| DE | 144 | 1182 | 40 | 2 | 90 | 275 | 19 | 5 | |
| DD|DE | 23 | 162 | 14 | 1 | 4 | 13 | 4 | 0 | |
| Mechanism | M0 | 3692 | 1177 | 3851 | 602 | 2565 | 1963 | 526 | 2334 |
| MI | 164 | 613 | 69 | 47 | 107 | 315 | 7 | 56 | |
| MD | 55 | 57 | 41 | 36 | 34 | 60 | 9 | 60 | |
| MM | 64 | 880 | 31 | 3 | 90 | 155 | 14 | 16 | |
| MT | 9 | 10 | 4 | 14 | 23 | 0 | 1 | 13 | |
| MS | 1 | 2 | 15 | 33 | 3 | 2 | 0 | 38 | |
| MN | 1 | 4 | 4 | 0 | 0 | 3 | 0 | 1 | |
| MA | 2 | 1 | 10 | 1 | 2 | 0 | 0 | 4 | |
| MD|MM | 2 | 12 | 0 | 0 | 6 | 1 | 0 | 1 | |
| MD|MT | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| MD|MI | 12 | 17 | 1 | 10 | 5 | 18 | 2 | 4 | |
| MD|MN | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MS | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| MI|MS | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 1 | |
| MI|MN | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| MI|MT | 4 | 5 | 3 | 1 | 6 | 0 | 2 | 0 | |
| MI|MM | 2 | 391 | 8 | 1 | 28 | 23 | 3 | 0 | |
| MM|MT | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MM|MN | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| MA|MS | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MA|MI | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| MS|MT | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Corpus . | DDI . | Non-DDI . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study categories . | In vivo DDI PK . | In vitro DDI PK . | DDI Clinical . | In vitro PD . | Drug–Nutrition . | Single Drug . | Case Reports . | Nondrug Studies . | |
| Focus | G | 226 | 144 | 272 | 110 | 263 | 253 | 68 | 306 |
| M | 976 | 218 | 944 | 80 | 664 | 374 | 46 | 382 | |
| S | 2740 | 2670 | 2725 | 531 | 1861 | 1785 | 440 | 1727 | |
| S|M | 65 | 141 | 90 | 38 | 90 | 127 | 10 | 120 | |
| S|G | 486 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | |
| G|M | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 3 | |
| Polarity | P | 3325 | 2868 | 3510 | 727 | 2481 | 2217 | 502 | 2319 |
| N | 685 | 308 | 532 | 33 | 400 | 323 | 62 | 219 | |
| Certainty | 0 | 340 | 315 | 373 | 71 | 244 | 220 | 9 | 198 |
| 1 | 20 | 31 | 17 | 1 | 9 | 11 | 3 | 5 | |
| 2 | 319 | 319 | 288 | 76 | 187 | 219 | 85 | 280 | |
| 3 | 3331 | 2521 | 3364 | 612 | 2441 | 2090 | 467 | 2055 | |
| Evidence | E0 | 319 | 172 | 209 | 39 | 103 | 140 | 61 | 232 |
| E1 | 214 | 197 | 276 | 123 | 113 | 127 | 55 | 202 | |
| E2 | 1 | 2 | 1 | 2 | 0 | 2 | 0 | 3 | |
| E3 | 2017 | 2028 | 2195 | 553 | 1669 | 1551 | 350 | 1567 | |
| EN | 1371 | 777 | 1361 | 43 | 996 | 720 | 98 | 534 | |
| Direction | + | 488 | 194 | 607 | 121 | 379 | 258 | 41 | 278 |
| − | 389 | 185 | 359 | 75 | 313 | 248 | 59 | 163 | |
| [+|−] | 2 | 2 | 14 | 0 | 1 | 4 | 0 | 1 | |
| Study Type | VV | 1818 | 93 | 159 | 10 | 996 | 359 | 54 | 14 |
| VT | 49 | 1499 | 22 | 396 | 163 | 732 | 0 | 133 | |
| VC | 300 | 46 | 2620 | 17 | 440 | 534 | 279 | 584 | |
| V0 | 1673 | 1417 | 1097 | 233 | 1125 | 852 | 196 | 585 | |
| VN | 92 | 67 | 77 | 97 | 54 | 27 | 29 | 1203 | |
| VV|VC | 72 | 5 | 64 | 0 | 95 | 21 | 5 | 1 | |
| VV|VT | 5 | 46 | 2 | 6 | 6 | 15 | 1 | 16 | |
| VT|VC | 1 | 3 | 0 | 1 | 2 | 0 | 0 | 2 | |
| Interaction | D0 | 841 | 548 | 1269 | 291 | 1048 | 953 | 174 | 2327 |
| Type | DR | 1131 | 857 | 1321 | 279 | 1584 | 1265 | 218 | 201 |
| DD | 1871 | 527 | 1398 | 187 | 155 | 25 | 149 | 5 | |
| DE | 144 | 1182 | 40 | 2 | 90 | 275 | 19 | 5 | |
| DD|DE | 23 | 162 | 14 | 1 | 4 | 13 | 4 | 0 | |
| Mechanism | M0 | 3692 | 1177 | 3851 | 602 | 2565 | 1963 | 526 | 2334 |
| MI | 164 | 613 | 69 | 47 | 107 | 315 | 7 | 56 | |
| MD | 55 | 57 | 41 | 36 | 34 | 60 | 9 | 60 | |
| MM | 64 | 880 | 31 | 3 | 90 | 155 | 14 | 16 | |
| MT | 9 | 10 | 4 | 14 | 23 | 0 | 1 | 13 | |
| MS | 1 | 2 | 15 | 33 | 3 | 2 | 0 | 38 | |
| MN | 1 | 4 | 4 | 0 | 0 | 3 | 0 | 1 | |
| MA | 2 | 1 | 10 | 1 | 2 | 0 | 0 | 4 | |
| MD|MM | 2 | 12 | 0 | 0 | 6 | 1 | 0 | 1 | |
| MD|MT | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| MD|MI | 12 | 17 | 1 | 10 | 5 | 18 | 2 | 4 | |
| MD|MN | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| MD|MS | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| MI|MS | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 1 | |
| MI|MN | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| MI|MT | 4 | 5 | 3 | 1 | 6 | 0 | 2 | 0 | |
| MI|MM | 2 | 391 | 8 | 1 | 28 | 23 | 3 | 0 | |
| MM|MT | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MM|MN | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| MA|MS | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | |
| MA|MI | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| MS|MT | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
Inter-annotator agreement on sentence fragmentation
IAA was first assessed for the sentence-fragmentation task. IAA in fragmentation between any pair of annotators among all abstracts was calculated using Equation (2). In the first round of annotation, IAA was 0.81, and it increased to 0.92 in the second round (P < 0.001). Figure 3 shows statistically significant improvement in fragmentation agreement for the following abstract subcategories: PK DDI in vivo, PK DDI in vitro, DDI clinical, PD DDI in vitro, single drug and nondrug (P < 0.05). Although the improvement in fragmentation agreement was not statistically significant for drug–nutrition interactions and case reports, their agreements all exceed 0.91 in the second round of annotations. The fragmentation agreement between any two experienced annotators (EE) (0.92, P = 0.003) was higher than that between any novice annotator and any experienced annotator (NE) (0.85) in the first round. After training, the agreement between two novice annotators improved to 0.92 in the second round.
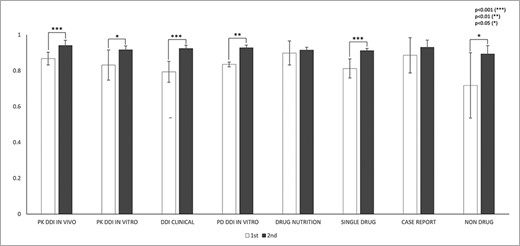
IAAs in fragmentation for eight abstract subcategories. This figure shows the IAAs (mean ± SEM) of two round annotations for fragmentation across eight abstract subcategories. The IAAs of Round 1 are shown as white bars and IAAs of Round 2 are black bars. The asterisk brackets added above the bars indicate statistically significant differences. Error bar represents the standard error of mean. The x-axis labels the eight annotation dimensions, and y-axis represents the IAA.
Annotation agreement over the eight annotation dimensions
The IAA for the eight annotation dimensions was assessed based on the mutually agreed fragmentations. There were totally 14 770 mutually agreed fragments in the first round and 16 142 fragments in the second round. Figure 4 and Table 5 show the IAA during two rounds of annotations. In general, IAA was higher in the second round across all eight dimensions. The agreement improved significantly in seven dimensions: focus (P < 0.001), polarity (P < 0.05), certainty (P < 0.001), study type (P < 0.001), interaction type (P < 0.001) and mechanisms (P < 0.001). Although the agreement increase in direction was not statistically significant (P = 0.053) from the first to the second round, the agreement was already very high.
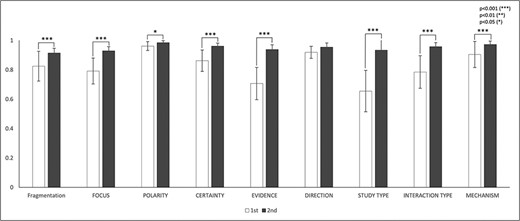
IAAs in eight annotation dimensions. This figure shows the IAAs (mean ± SEM) for eight dimensions in Round 1 and 2 annotations. The IAAs of Round 1 are shown in white bars and the agreements of Round 2 are black bars. The asterisk brackets added above the bars indicate statistically significant differences. Error bar represents the standard error of mean. The x-axis labels the eight annotation dimensions, and y-axis represents the IAA.
| IAA . | First round . | Second round . |
|---|---|---|
| Fragmentation | 0.82 | 0.91 |
| Focus | 0.79 | 0.93 |
| Polarity | 0.96 | 0.98 |
| Certainty | 0.86 | 0.96 |
| Evidence | 0.71 | 0.94 |
| Direction | 0.92 | 0.95 |
| Study Type | 0.65 | 0.93 |
| Interaction Type | 0.78 | 0.96 |
| Mechanism | 0.9 | 0.97 |
| IAA . | First round . | Second round . |
|---|---|---|
| Fragmentation | 0.82 | 0.91 |
| Focus | 0.79 | 0.93 |
| Polarity | 0.96 | 0.98 |
| Certainty | 0.86 | 0.96 |
| Evidence | 0.71 | 0.94 |
| Direction | 0.92 | 0.95 |
| Study Type | 0.65 | 0.93 |
| Interaction Type | 0.78 | 0.96 |
| Mechanism | 0.9 | 0.97 |
| IAA . | First round . | Second round . |
|---|---|---|
| Fragmentation | 0.82 | 0.91 |
| Focus | 0.79 | 0.93 |
| Polarity | 0.96 | 0.98 |
| Certainty | 0.86 | 0.96 |
| Evidence | 0.71 | 0.94 |
| Direction | 0.92 | 0.95 |
| Study Type | 0.65 | 0.93 |
| Interaction Type | 0.78 | 0.96 |
| Mechanism | 0.9 | 0.97 |
| IAA . | First round . | Second round . |
|---|---|---|
| Fragmentation | 0.82 | 0.91 |
| Focus | 0.79 | 0.93 |
| Polarity | 0.96 | 0.98 |
| Certainty | 0.86 | 0.96 |
| Evidence | 0.71 | 0.94 |
| Direction | 0.92 | 0.95 |
| Study Type | 0.65 | 0.93 |
| Interaction Type | 0.78 | 0.96 |
| Mechanism | 0.9 | 0.97 |
Figure 5 shows in-depth IAA analysis for each of eight annotation dimensions among DDI and non-DDI abstract subcategories: PK DDI in vivo, PK DDI in vitro, clinical DDI, PD DDI in vitro, drug–nutrition, single drug, case reports and nondrug. IAAs in Evidence (Figure 5D) and Study Type (Figure 5F) were universally improved from the first round to the second round among all eight abstract subcategories (P < 0.05). Polarity (Figure 5B) and Direction (Figure 5E) showed IAA improvement in only two out of eight abstract subcategories (P < 0.05) from the first to the second round, because their first-round IAAs are relatively high already. The other dimensions, such as Focus (Figure 5A), Certainty (Figure 5C), Interaction Type (Figure 5G) and Mechanisms (Figure 5H), showed IAA improvement in some but not all abstract subcategories (P < 0.05).
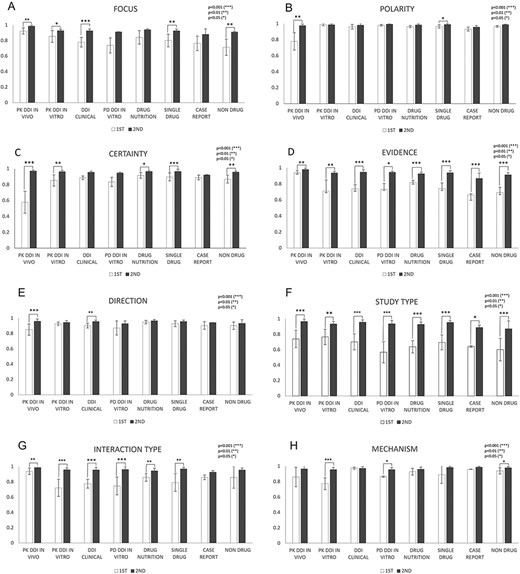
IAAs in eight annotation dimensions among different abstract subcategories. This figure shows IAAs (mean ± SEM) for each dimension in different abstract subcategories. The IAAs of Round 1 are shown as white bars and the IAAs of Round 2 are black bars. The asterisk brackets added above the bars indicate statistically significant differences. Error bar represents the standard error of mean. The x-axis labels the eight annotation dimensions, and y-axis represents the IAA.
Inter-annotator agreements between experienced and novice annotators
Figure 6 illustrates IAAs between novice and experienced annotators (NE) and between two experienced annotators (EE) in both rounds. Overall, IAAs universally improved from Rounds 1 and 2 among NE and EE annotator pairs in fragmentation and eight annotation dimensions. In Round 1, IAAs of EE were higher than those of NEs in five annotation dimensions, namely, Certainty (P < 0.001), Evidence (P < 0.01), Study Type (P < 0.01), Interaction Type (P < 0.001) and Mechanism (P < 0.01). For the other three dimensions: Focus, Polarity and Direction, EEs had slightly higher IAAs than NEs, although not statistically significant. In particular, NEs had very low IAAs in Evidence (0.73) and Study Type (0.72) during the first round of annotation. In Round 2, NEs had IAAs all improved such that their IAAs became almost indistinguishable from EE IAAs.
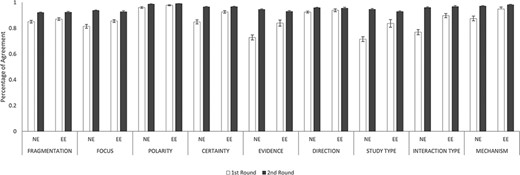
IAAs between novice and experienced annotators and between two experienced annotators. This figure shows the IAAs (mean ± SEM) between novice and experienced annotators (NE) and between two experienced annotators (EE), in fragmentation and eight annotation dimensions. The IAAs of Round 1 are shown as white bars and Round 2 IAAs are black bars. Error bar represents standard error of mean. The x-axis labels the eight annotation dimensions, and y-axis represents the IAA.
Discussion and conclusion
Our DDI corpus comprises 900 DDI and 750 non-DDI abstracts. Each abstract was first broken into text fragments, and each text fragment was then further annotated along eight dimensions, which included study type, interaction type and DDI mechanism as well as focus, polarity, certainty, evidence and directionality. Unlike earlier DDI corpus, our new corpus used sentence fragments as its basic annotation unit. We also separated our corpus into eight categories of DDI or non-DDI abstracts, namely in vitro DDI PK, in vivo DDI PK, DDI clinical, drug–nutrition interaction, single drug, no drug related, in vitro PD and case report. Although previous corpora categorized abstracts into DDI and PK categories (18–22), the abstracts included in our corpus are further categorized, for the first time, into finer categories, namely, drug–nutrition, no drug, in vitro PD and case report. In addition, our corpus further adds focus, polarity, certainty, evidence and directionality.
The most important contribution of our corpus is differentiated DDI evidence in vitro DDI PK, in vivo DDI PK and clinical DDI. This will allow future translational DDI knowledge gap discovery research, using machine learning and artificial intelligence.
To develop a high-quality corpus, seven annotators, including three experienced annotators and four novice annotators, conducted two rounds of annotations. Novice annotators received additional training after the first round of annotation. Then, in the second round of annotation, the novice annotators performed as well as the experienced annotators. Agreement in the second round of annotations was close between NE annotator pairs and EE annotator pairs. IAAs significantly improved in all eight annotation dimensions from the first round to the second round, and agreement in all dimensions exceeded 92% after the second round.
In reviewing fragment annotation frequencies reported from different abstract types, we notice that some fragments or sentences in non-DDI abstracts contain evidence of drug interaction, such as DDI and drug–enzyme interaction. This creates an additional layer of complexity and a challenge in the follow-up retrieval of drug–interaction-related abstracts.
Our DDI corpus can be used by the BioNLP community and promote the development of text-mining techniques for the detection of DDI in biomedical text. The corpus described in this work, including both the annotated corpus and the annotation guidelines, are available at https://github.com/zha204/DDI-Corpus-Database/tree/master/DDI%20corpus.
Funding
This work was supported by the National Institutes of Health grants (R01LM011945).
Conflict of interest
None declared.
Data availability
This database is freely available at GitHub.
References
Author notes
contributed equally to this work.



