-
PDF
- Split View
-
Views
-
Cite
Cite
Tiago Almeida, Rui Antunes, João F. Silva, João R Almeida, Sérgio Matos, Chemical identification and indexing in PubMed full-text articles using deep learning and heuristics, Database, Volume 2022, 2022, baac047, https://doi.org/10.1093/database/baac047
Close - Share Icon Share
Abstract
The identification of chemicals in articles has attracted a large interest in the biomedical scientific community, given its importance in drug development research. Most of previous research have focused on PubMed abstracts, and further investigation using full-text documents is required because these contain additional valuable information that must be explored. The manual expert task of indexing Medical Subject Headings (MeSH) terms to these articles later helps researchers find the most relevant publications for their ongoing work. The BioCreative VII NLM-Chem track fostered the development of systems for chemical identification and indexing in PubMed full-text articles. Chemical identification consisted in identifying the chemical mentions and linking these to unique MeSH identifiers. This manuscript describes our participation system and the post-challenge improvements we made. We propose a three-stage pipeline that individually performs chemical mention detection, entity normalization and indexing. Regarding chemical identification, we adopted a deep-learning solution that utilizes the PubMedBERT contextualized embeddings followed by a multilayer perceptron and a conditional random field tagging layer. For the normalization approach, we use a sieve-based dictionary filtering followed by a deep-learning similarity search strategy. Finally, for the indexing we developed rules for identifying the more relevant MeSH codes for each article. During the challenge, our system obtained the best official results in the normalization and indexing tasks despite the lower performance in the chemical mention recognition task. In a post-contest phase we boosted our results by improving our named entity recognition model with additional techniques. The final system achieved 0.8731, 0.8275 and 0.4849 in the chemical identification, normalization and indexing tasks, respectively. The code to reproduce our experiments and run the pipeline is publicly available.
Database URL
Introduction
Over the years, researchers have published scientific articles in various venues such as journals, conferences and, more recently, public open archives. With the increasing pace of research, an enormous amount of new articles are now published on a daily basis. For instance, in 2020 PubMed alone was responsible for the indexing of ∼1.5 million new articles (https://www.nlm.nih.gov/bsd/medline_pubmed_production_stats.html), which corresponds to a publishing rate of nearly three new articles every minute. This rapid literature generation rate becomes, as noted by Landhuis et al. (1), a burden to researchers who need to invest more time for keeping track of current literature while continuing with their ongoing research work.
Automatic information extraction systems are viewed as possible solutions to aid researchers in tasks that deal with overwhelming volumes of data, as suggested by Grishman (2). Information extraction can (i) help researchers to quickly grasp the knowledge encoded in each scientific article; (ii) help data curators to expedite their work or (iii) help automatic search systems to improve their performance by directly indexing extracted terms from scientific manuscripts. Specifically, the information extraction task of named entity recognition (NER) focuses on directly identifying entities in free text, such as names of diseases, chemicals or genes. Of particular interest is the identification of chemical names, which are among the most frequently searched entity types in PubMed (3), given the potential impact on tasks such as drug–drug interaction extraction and detection of adverse drug events and finally on drug development. In this context, it is also usual to normalize the identified chemical mentions by linking them to unique codes from a standard vocabulary, such as Medical Subject Headings (MeSH) (4).
Despite the added value of using the extra information present in PubMed full-text articles, biomedical information extraction systems have typically limited their scope to PubMed abstracts owing, on one hand, to the open availability, and on the other, to the challenges stemming from the more complex writing style of the full-text content, which contain more detailed explanations and statements when compared to abstracts. Herein we propose an end-to-end processing system for the extraction, normalization and indexing of chemical-related entities, operating over full-text articles. The system was designed as a cascaded pipeline that addresses each task in an isolated manner, containing the following three core modules: ‘Annotator’, ‘Normalizer’ and ‘Indexer’. Firstly, the ‘Annotator’ module aims to identify chemical entities in full-text articles, leveraging current advances in deep learning to create a neural model capable of recognizing chemical entities. Next, the ‘Normalizer’ uses a rule-based method based on dictionary lookup combined with a deep-learning-based approach for linking previously identified entities to their corresponding MeSH codes. Finally, the ‘Indexer’ explores a rule-based method and a term frequency–inverse document frequency (TF-IDF) approach for the selection of the most relevant MeSH codes to be indexed per article.
Our system was originally developed and evaluated within the context of the BioCreative VII Track 2 (NLM-Chem) challenge (5), which aimed to bring together the text-mining community in order to improve the state of the art in chemical entity identification. The NLM-Chem challenge track was divided into two tasks: (i) chemical identification and (ii) chemical indexing. In the first task, the objective was to recognize chemical mentions (NER) in full-text scientific articles and link the predicted entities to their corresponding MeSH identifiers, also known as normalization. The second task aimed to predict the chemical MeSH identifiers that should be used to index each document, i.e., find the most relevant MeSH terms for each document. The proposed system was further improved in a post-challenge contribution, with higher emphasis being given to the NER module since it is the first piece of the end-to-end pipeline where improvements at the beginning of the pipeline cascade to the downstream modules.
The present paper is structured as follows: in ‘Related work’ we contextualize our work with the current literature; in ‘Materials and methods’ we address all of the resources used in this work and provide a complete description of the proposed end-to-end pipeline, detailing the methods developed within each module as well as post-challenge improvements; in the ‘Results and discussion’ section we show the results obtained in the BioCreative VII Track 2 challenge along with improved results obtained post-challenge and present important insights together with some ablation studies that supported the choice of our methods; finally, in ‘Conclusions and future work’ we provide other existing possibilities for further improving the proposed solution at the three different levels of the pipeline.
Related work
The use of computerized solutions to ease the work of biological, clinical and medical researchers has been a field of growing investigation during the past decades (6, 7). In the life sciences field, information access platforms such as the PubMed Central (8) that are capable of retrieving pertinent documents according to the information needs of researchers are important to expedite and facilitate their investigations. Articles linked in PubMed are indexed by expert curators that use MeSH code identifiers to attribute concepts of interest to each document. Information extraction systems have also been intensively researched and developed (9), allowing the automatic mining of key knowledge that helps to keep biomedical databases updated and alleviating the need for manual efforts (10, 11). For instance, previous research efforts on information extraction have focused on identifying biomedical entities such as genes, proteins, chemical compounds (12) and clinical entities including laboratory procedures, diseases and adverse effects (13, 14). Identification of these entities of interest in the text is commonly paired with a normalization step, where the entities are grounded to unique identifiers from standard vocabularies or databases. This avoids ambiguity which is in general accentuated in the biomedical domain (15), and it is a recurrent obstacle for text-mining methods. For example, genes and chemicals are commonly mapped using the Entrez Gene and ChEBI databases, respectively (16, 17). A further step of extraction using the identified entities is about extracting interactions between the mentions of interest. Past research work has addressed extracting gene–disease (18), protein–protein (19, 20), chemical–disease (21) and chemical–protein relations (22, 23).
In this section we focus on reviewing past literature about the research problems that are addressed in this manuscript, including NER, entity normalization and indexing. The recognition of named entities is considered a key component for information extraction (2), serving several purposes such as a lever for relation extraction (24), entity linking (25) and other text-mining purposes. NER is commonly treated as a sequence labeling problem, where (sub-)words need to be classified as a part of entity or not. For that purpose, the BIO (beginning, inside, outside) tagging schema is the most common option for token-level classification and has been extensively explored (26–28). Campos et al. (29) make an extensive survey of machine learning tools in biomedical NER. Habibi et al. (30) used a long short-term memory–conditional random field (LSTM-CRF) architecture using ‘word2vec’ word embeddings (31) pre-trained on biomedical corpora (32) and evaluated their model on 33 datasets covering different entity types. Lample et al. (28) propose the use of bidirectional LSTMs (BiLSTMs) and CRFs, showing its effectiveness in deep-learning-based approaches for NER. More recent works have been addressing the NER task jointly with other tasks. Particularly, the joint extraction of entities and their relations has been a growing area of research. Major works, just to name a few, include the proposal of an end-to-end neural model that uses LSTMs on sequence and tree structures (33) and the multi-head selection problem tackled with a BiLSTM-CRF model (34).
Entity normalization, linking or grounding, is either followed by NER in a pipeline fashion, or these two tasks can be jointly addressed in a multitask learning setup. In the biomedical domain, two of the most known tools for concept normalization are MetaMap (35, 36) and cTAKES (37) which are mainly based on dictionary lookup techniques. Leaman et al. (38) addressed the normalization of disease mentions in PubMed abstracts using a machine learning approach, which was evaluated on the NCBI disease corpus (39). The SemEval 2015 competition (40) similarly addressed the task of disorder identification. For instance, one of the participating teams, Leal et al. (41), presented a chain of two modules: one for recognition based on CRFs and another for normalization based on dictionaries and heuristics. This approach is similar to our end-to-end system described in this manuscript, since we also use a CRF for mention recognition and dictionaries for entity linking. Leaman et al. (42) also used CRF models in the tmChem system for chemical identification and normalization, which combines two independent machine learning in an ensemble for recognizing the chemicals, whereas a simple lexical approach is employed for normalization. Leaman et al. (43) further investigated the application of machine learning models for disease normalization in clinical narratives, since previous research had shown deteriorated performance in comparison to biomedical scientific publications. Also, Leaman and Lu (44) proposed the first machine learning model that jointly tackled NER and normalization during training and inference and released the TaggerOne toolkit that can be applied to any entity type. The authors assessed their system in the NCBI disease (39) and in the CDR (45, 46) corpora. Perez-Miguel et al. (47) used the Unified Medical Language System (UMLS) Metatheusaurus for term normalization in clinical Spanish text. Luo et al. (48) created the Medical Concept Normalization corpus which targeted normalization of medical concepts found in the clinical free-text notes of electronic health records and was used during the 2019 n2c2/UMass Lowell shared task (49). Our research group also participated in this challenge (50) employing the BioWordVec model (51) for representing clinical terms and showed the effectiveness of distributed word representations in entity normalization. Zhao et al. (52) proposed a deep neural multitask learning model, with explicit feedback strategies, to jointly tackle medical NER and normalization, by (i) using the same representations for both tasks and (ii) using a parallel setup while maintaining the mutual support between them. The authors used BiLSTMs to improve the sequential modeling of text and convolutional neural networks (CNNs) to detect character-level clues such as Zolmitriptan, Zomig and Zomigon. Similarly, Kim et al. (53) present a neural NER and normalization tool for biomedical text, particularly addressing the problem of overlapping entities which is frequently observed in text that is annotated with entities of multiple types (chemicals, genes, diseases and others). They developed probability-based decision rules to identify the types of overlapping entities and integrated various NER models, which helped in assigning a distinct identifier to each recognized entity. Their tool, named BERN, made use of BioBERT (54) NER models to improve the discovery of new entities. More recently, Luo et al. (55) developed the package pyMeSHSim for recognizing and normalizing biomedical named entities using MetaMap (36). Xu et al. (56) designed a sieve-based system based on Apache Lucene indices over training data and collected information from UMLS resources to generate a list of candidate concepts for each recognized entity. They then applied a listwise classifier based on the BioBERT neural network (54) to rank the possible identifiers. Ruas et al. (57) developed a system for normalization of tumor morphology entities in Spanish health-related documents under the participation for the CANTEMIST competition (58). The authors used a BiLSTM-CRF tagger for NER and adopted a graph-based model to rank concept candidates for each entity mention. Many other recent works have also tackled hybrid approaches (59) and edit patterns (60), analyzed the problem of ambiguity (61), explored transformer networks trained via a triplet objective (62) and multi-task frameworks (63) and experimented using large-scale datasets (64).
Early efforts in MeSH indexing include the ‘Gene Indexing initiative’ by the National Library of Medicine, which was analyzed by Mitchell et al. (65) concluding that it was helpful for the life sciences research community. Since then, several methods for automatic indexing in biomedical scientific literature have been proposed. Jimeno-Yepes et al. (66) compared and combined several MeSH indexing approaches. Liu et al. (67) proposed the MeSHLabeler framework that integrates multiple evidence from machine learning classifiers, pattern matching and other predictions. Peng et al. (68) proposed DeepMeSH, making use of sparse and dense semantic representations. Irwin and Rackham (69) performed an extensive study about the time-to-indexing PubMed documents in different journals. Mao and Lu (70) proposed MeSH Now that first ranks the candidate identifiers and then uses a post-processing module to select the highest-ranked MeSH terms. Dai et al. (71) proposed FullMeSH, which makes use of full-text and gives distinct importance to different sections of the article. You et al. (72) introduced BERTMeSH, a full-text and deep-learning-based MeSH indexing method. Costa et al. (73) explored MeSH headings to index health news. Finally, Rae et al. (74) present a new neural text ranking approach based on PubMedBERT for automatic MeSH indexing.
Materials and methods
This section introduces the data used for training and evaluating our system, specifies the adopted evaluation metrics and lastly provides a detailed description on the development of the three core modules of the end-to-end pipeline.
Data
The data used in this work—NLM-Chem BioCreative VII corpus—was developed for chemical identification and indexing in full-text articles and consisted in three parts as explained in detail by Islamaj et al. (75):
The NLM-Chem200 corpus contains 204 full-text PubMed articles that were manually annotated with chemical entities (both mention boundaries and normalization identifiers) and indexing codes. The training set consists of 150 documents, whereas the remaining 54 documents were used for official evaluation of the chemical identification task;
Extended collection from previous BioCreative challenges comprising 11 500 PubMed abstracts from the CDR (46) and CHEMDNER (76) datasets from past BioCreative editions, which were enriched with chemical indexing codes;
The chemical indexing testing dataset was composed of 1 387 PubMed articles annotated with indexed MeSH codes. Since this dataset had a significantly higher number of documents compared to the 54 articles used to evaluate the chemical identification task, it enabled a more solid evaluation of indexing solutions and hindered manual annotation by the participants during the challenge.
The NLM-Chem dataset was composed of two parts: a first one for system development (training set) and another for official evaluation of challenge submissions (final test set). The training set was additionally partitioned by the organizers in three subsets: ‘train’, ‘dev’ and ‘test’. The NLM-Chem final test set differed according to the task under evaluation. The ‘Chemical Identification’ task used a NER test set containing 54 documents with gold-standard chemical annotations, whereas for ‘Chemical Indexing’ a test set containing 1387 documents with indexed MeSH codes was employed.
Task organizers provided additional datasets to encourage teams to use more training data: (i) the CDR corpus (46) was used in the BioCreative V challenge for text mining on chemical–disease relations and (ii) the CHEMDNER corpus (76) was employed in BioCreative IV for chemical mention recognition. Although both corpora contain PubMed abstracts annotated with chemical entities and MeSH indexing codes, only the CDR corpus has entities linked to MeSH codes for normalization.
To train our NER deep-learning model we used all the datasets made available by the organizers. Furthermore, we explored other chemical-related datasets during initial experiments for the official submissions for BioCreative challenge, namely the CRAFT, BioNLP11ID, BioNLP13CG and BioNLP13PC datasets prepared by Crichton et al. (77). However, due to computational limitations and the lack of significant benefits from using these datasets, we opted to drop them in the post-challenge experiments described within this manuscript. For further details about the use of these datasets in our former experiments we point the reader to the proceedings paper describing the system used in official challenge submissions (78). Also, we note that these datasets were not annotated following the CHEMDNER annotation guidelines (76), which could deteriorate NER performance on the NLM-Chem dataset. In contrast, chemicals in the CDR and CHEMDNER corpora were annotated using the CHEMDNER guidelines, and NLM-Chem was similarly annotated using specific guidelines (79) that were based on these. The BioCreative VII edition also held the DrugProt track (23), which addressed the extraction of relationships between drugs (chemical compounds) and proteins (genes). We also explored the DrugProt dataset since it contained gold-standard chemical entities that were annotated following the CHEMDNER guidelines.
Detailed statistics on the aforementioned corpora are presented in Table 1. Regarding the NLM-Chem corpus, the final NER test set contained approximately twice the number of chemical mentions present in ‘test’ subset of the training set. We hypothesize this may be due to the fact that the selected articles contained more chemicals or that text from these articles was overall longer. As expected, it is also noticeable that corpora containing only abstracts have a few number of chemicals per document (∼ten chemicals per abstract) in comparison to the full-text NLM-Chem corpus (around 200–400 chemicals per document). Regarding indexing annotations, the NLM-Chem training set has an average of 2.43 MeSH indexed codes per document, whereas the NLM-Chem final test set has an average of 2.87 MeSH indexed codes. We suspect that this increase in the number of MeSH indexing identifiers in the final test set (around 18%) can be justified in part by the higher number of chemicals present in the documents.
Dataset statistics with the number of documents, chemicals and MeSH indexing identifiers. PMID: PubMed identifier.
| . | Documents . | Chemical mentions . | Indexing identifiers . |
|---|---|---|---|
| NLM-Chem | |||
| Training set (total) | 150 | 38 339 | 364 |
| ‘train’ subset | 80 | 21 218 | 204 |
| ‘dev’ subset | 20 | 5349 | 51 |
| ‘test’ subset | 50 | 11 772 | 109 |
| Final evaluation set | |||
| NER test set | 54 | 22 942 | – |
| Indexing test set | 1387 | – | 3980 |
| CDR (total) | 1500 | 15 943 | 3461 |
| ‘train’ subset | 500 | 5205 | 1166 |
| ‘dev’ subset | 500 | 5349 | 1198 |
| ‘test’ subset | 500 | 5389 | 1097 |
| CHEMDNER (total) | 10 000 | 84 331 | 21 565 |
| ‘train’ subset | 3500 | 29 462 | 7692 |
| ‘dev’ subset | 3500 | 29 523 | 7666 |
| ‘test’ subset | 3000 | 25 346 | 6207 |
| DrugProta (total) | 2180 | 33 866 | – |
| ‘train’ subset | 1781 | 27 720 | – |
| ‘dev’ subset | 399 | 6146 | – |
| . | Documents . | Chemical mentions . | Indexing identifiers . |
|---|---|---|---|
| NLM-Chem | |||
| Training set (total) | 150 | 38 339 | 364 |
| ‘train’ subset | 80 | 21 218 | 204 |
| ‘dev’ subset | 20 | 5349 | 51 |
| ‘test’ subset | 50 | 11 772 | 109 |
| Final evaluation set | |||
| NER test set | 54 | 22 942 | – |
| Indexing test set | 1387 | – | 3980 |
| CDR (total) | 1500 | 15 943 | 3461 |
| ‘train’ subset | 500 | 5205 | 1166 |
| ‘dev’ subset | 500 | 5349 | 1198 |
| ‘test’ subset | 500 | 5389 | 1097 |
| CHEMDNER (total) | 10 000 | 84 331 | 21 565 |
| ‘train’ subset | 3500 | 29 462 | 7692 |
| ‘dev’ subset | 3500 | 29 523 | 7666 |
| ‘test’ subset | 3000 | 25 346 | 6207 |
| DrugProta (total) | 2180 | 33 866 | – |
| ‘train’ subset | 1781 | 27 720 | – |
| ‘dev’ subset | 399 | 6146 | – |
a The DrugProt dataset was filtered to discard repeated documents (sharing the same PMID) already annotated in other corpora. The test set partition of DrugProt was not used because at the time of experiments the DrugProt organizers did not release it to the public domain.
Dataset statistics with the number of documents, chemicals and MeSH indexing identifiers. PMID: PubMed identifier.
| . | Documents . | Chemical mentions . | Indexing identifiers . |
|---|---|---|---|
| NLM-Chem | |||
| Training set (total) | 150 | 38 339 | 364 |
| ‘train’ subset | 80 | 21 218 | 204 |
| ‘dev’ subset | 20 | 5349 | 51 |
| ‘test’ subset | 50 | 11 772 | 109 |
| Final evaluation set | |||
| NER test set | 54 | 22 942 | – |
| Indexing test set | 1387 | – | 3980 |
| CDR (total) | 1500 | 15 943 | 3461 |
| ‘train’ subset | 500 | 5205 | 1166 |
| ‘dev’ subset | 500 | 5349 | 1198 |
| ‘test’ subset | 500 | 5389 | 1097 |
| CHEMDNER (total) | 10 000 | 84 331 | 21 565 |
| ‘train’ subset | 3500 | 29 462 | 7692 |
| ‘dev’ subset | 3500 | 29 523 | 7666 |
| ‘test’ subset | 3000 | 25 346 | 6207 |
| DrugProta (total) | 2180 | 33 866 | – |
| ‘train’ subset | 1781 | 27 720 | – |
| ‘dev’ subset | 399 | 6146 | – |
| . | Documents . | Chemical mentions . | Indexing identifiers . |
|---|---|---|---|
| NLM-Chem | |||
| Training set (total) | 150 | 38 339 | 364 |
| ‘train’ subset | 80 | 21 218 | 204 |
| ‘dev’ subset | 20 | 5349 | 51 |
| ‘test’ subset | 50 | 11 772 | 109 |
| Final evaluation set | |||
| NER test set | 54 | 22 942 | – |
| Indexing test set | 1387 | – | 3980 |
| CDR (total) | 1500 | 15 943 | 3461 |
| ‘train’ subset | 500 | 5205 | 1166 |
| ‘dev’ subset | 500 | 5349 | 1198 |
| ‘test’ subset | 500 | 5389 | 1097 |
| CHEMDNER (total) | 10 000 | 84 331 | 21 565 |
| ‘train’ subset | 3500 | 29 462 | 7692 |
| ‘dev’ subset | 3500 | 29 523 | 7666 |
| ‘test’ subset | 3000 | 25 346 | 6207 |
| DrugProta (total) | 2180 | 33 866 | – |
| ‘train’ subset | 1781 | 27 720 | – |
| ‘dev’ subset | 399 | 6146 | – |
a The DrugProt dataset was filtered to discard repeated documents (sharing the same PMID) already annotated in other corpora. The test set partition of DrugProt was not used because at the time of experiments the DrugProt organizers did not release it to the public domain.
Finally, inspired by the work of Kim et al. (80), which obtained the highest official NER result, a synthetic dataset was generated from the training set of the NLM-Chem corpus, here denoted as NLM-Chem-Syn. Similarly, we employed their approach based on synonym replacement (81) which consisted in replacing chemical mentions by their synonyms present in the Comparative Toxicogenomics Database (82). We created several synthetic datasets using the different partitions of the NLM-Chem training set, with distinct sizes (1–4 times larger than the original set), and several values for the synonym replacement ratio (0.1, 0.3, 0.5 and 0.7), i.e. the probability of replacing a chemical mention with one of its synonyms. However, due to limitations in computational power we could not experiment every NLM-Chem-Syn variant. Instead, we empirically selected and experimented with only one synthetic dataset, using a dataset two times larger than the original one with a probability of 0.5 for synonym replacement.
Performance evaluation
The BioCreative VII NLM-Chem organizers considered the precision, recall and F1-score metrics for evaluation purposes, using the F1-score as the final metric to rank all participating teams in the three different tasks: entity recognition, entity normalization and MeSH code indexing. Challenge organizers defined two variants for each metric: a strict and an approximate version. Strict F1-score was used as the key ranking factor, whereas approximate F1-score was used to disambiguate rankings in the event of tied strict F1-scores.
Regarding the entity recognition subtask, a predicted chemical entity was considered a true positive (correct) if its boundaries matched the gold standard annotation, otherwise it was considered a false positive (incorrect). A false negative consisted in a case where the system failed to predict a gold-standard chemical annotation. To evaluate the system performance in the entity normalization subtask, the organizers only considered the set of distinct MeSH codes predicted for each document, disregarding the frequency of occurrence of each MeSH code (i.e. MeSH codes appearing once or multiple times within a document are attributed the same relevance in this evaluation process). Finally, similarly to the entity normalization subtask, indexing systems were evaluated considering the set of MeSH codes predicted to be indexed.
For simplicity purposes, in the development of our system we only used strict evaluation since it is simpler and more common, and its improvement is also reflected in the approximate evaluation metrics.
System pipeline
The conceptualization of our system was highly influenced by the BioCreative VII NLM-Chem track, where the organizers decided to split the problem into two main tasks: (i) ‘chemical identification’ and (ii) ‘chemical indexing’. However, the first task was further divided into two individual subtasks, (A) ‘chemical recognition’ and (B) ‘normalization’, leaving (C) ‘chemical indexing’ as a final task. Considering this division in three separate tasks, our system was developed as a three-stage cascade pipeline where in each stage we address each of the previous well-defined tasks. More precisely, in the first stage of the pipeline, the ‘Annotator’ module is focused on (A) chemical recognition and its respective annotation. The next piece in the pipeline is the ‘Normalizer’, which tackled the (B) chemical normalization task by linking each of the previously recognized entities to a standard code within the MeSH vocabulary. The final piece of this pipeline is the ‘Indexer’, whose objective is to select the most relevant chemical MeSH codes present in an article, thus addressing the (C) chemical indexing task.
Annotator
The ‘Annotator’ module has the objective of detecting the boundaries for chemical mentions in the raw document text. This task is well known in literature as NER and can be formulated as a sequence classification problem, where for each text token one must assign the most probable label according to the BIO (beginning, inside, other) tagging schema. Formally, let us consider |$\textbf{x}=\{x_1,x_2,{\ldots},x_N\}$| as a sequence of text tokens, where xi corresponds to the i-th token in the text and N to the total number of tokens (length of the text); |$\textbf{y}=\{y_1,y_2,{\ldots},y_N\}$| as a sequence of labels, where |$y_i\in \{B,I,O\}$| corresponds to the label of token xi; and |$P(\textbf{y}|\textbf{x})$| as the probability of a sequence of tokens x being labeled according to the sequence of labels y. Then, the objective is to find the most probable sequence of labels, |$\textbf{y*}$|, for any given input text, x, which mathematically corresponds to |$\textbf{y*}=\underset{\hat{\textbf{y}}\ \in\ \textbf{Y}}{argmax}\ P(\hat{\textbf{y}}|\textbf{x})$|, where |$\hat{\textbf{y}}$| represents a predicted label sequence and Y consists in the set containing all possible label sequences for x.
However, under the scope of this problem the independence assumption is flawed since there is a dependency between the entity token tags in the BIO tagging schema. For instance, after an O (Outside) tag it is impossible to have an I (Inside) tag, it must be a B (Beginning) or another O tag. Therefore, by also taking into consideration the previous label prediction, the model can more faithfully respect the BIO schema while gaining access to more context information when estimating a label probability yi given sequence x. An important note here is to distinguish between independent prediction and (in)dependent representations, as it is common to use LSTM networks or transformers to estimate |$P(y_i|\textbf{x})$|, which already imply that each token representation is contextualized by its surrounding tokens. However, this does not ensure that the prediction is dependent of the context (neighboring token labels), instead it ensures that the model has access to contextualized information to then make an independent prediction. Considering the aforementioned, Equation 1 was reformulated into Equation 2.
Here, |$P(y_i|\textbf{x})$| is the probability of a label yi being assigned to token xi given the complete sequence x, and |$P(y_i|y_{i-1})$| is the probability of yi being estimated given the previous predicted label, i.e. it accounts for the likelihood of a label being chosen given the previously predicted label. Fortunately, this is already a well-studied problem in literature, and if we consider the text as a simple directed graph, Equation 2 can be directly implemented as a linear-chain CRF, as described in Equation 3.
where θu and θt represent all trainable parameters that the model learns during the training phase, fu is the unary function parameterized by θu that computes the so-called unary potentials, i.e. it computes the score of each label being assigned to token xi while considering the whole sequence, and ft is the transition function that simply corresponds to a transition matrix, being parameterized by θt and having its score obtained by looking up the (|$y_{i-1},y_i$|) entry in the matrix. Lastly, |$Z(\textbf{x})$| is known as the partition function and acts as a normalizing factor to obtain a probabilistic distribution over all sequences.
To define fu we relied on the current state-of-the-art Bidirectional Encoder Representations from Transformers (BERT) model for creating contextualized word representations that are then forwarded through a multilayer perceptron (MLP) to compute the unary potentials. Particularly, we adopted the PubMedBERT (83) variant that reports the state-of-the-art results in almost every biomedical natural language processing task. As previously mentioned, this method produces contextualized unary potentials since the whole sentences are fed to PubMedBERT.
Due to the large size of BERT-based models (the base version has 110 million of parameters), these models cost a lot of time and computational resources to train and infer results. With this in mind, we devised a caching mechanism where it is possible to define the number of trainable layers from the BERT model, while the remaining ones become frozen and have their output stored in disk so that it can be reused during training. By leveraging this caching mechanism, we effectually produce a much lower memory footprint while massively speeding up training, at the cost of restricting the degrees of freedom from the model.
Furthermore, since we aimed to take full advantage of the contextualization power of the transformer architecture, we decided to set our input size to 512 tokens of which only the 256 tokens in the middle are forwarded to the MLP, whereas the remaining 256 tokens (128 to the left and 128 to the right) are only used for context. For visual reference, Figure 1 presents the full neural flow pipeline of the ‘Annotator’ module. Moreover, full-text documents from the NLM-Chem dataset are divided into passages, e.g. abstract, introduction, methods and others. Thus, each passage may comprise several sentences or paragraphs, which can surpass the BERT input limit of 512 tokens. To account for that, we split a passage into successive sequences shifted by 256 tokens (left-and-right context is kept), and each sequence is fed into PubMedBERT, as shown in Figure 2. An advantage of this method is that we can sequentially feed each passage of the document without performing any additional splitting such as sentence or paragraph segmentation. However, this method comes at a cost of computational time, since of the 512 tokens that are fed to the model, only the 256-centered are used for classification. Nevertheless, we employed this approach since it achieved the best preliminary results. Furthermore, we leave to future work to study the trade-off between the number of context tokens and tokens used for classification, since this can be viewed as an optimization step.
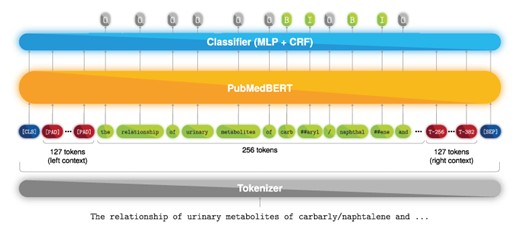
Named entity recognition deep learning model. Example text from PMC 1 253 656.
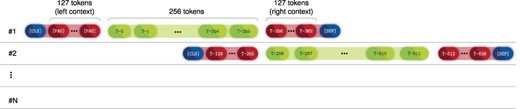
Diagram showing how token sequences are shifted in order to be fed into BERT.
For model training, we adopted the modern AdamW optimizer to minimize the negative log likelihood of Equation 3 and used the non-monotonic Mish activation function for the MLP. Additionally, we also experimented with training the last layer of the PubMedBERT model and using additional datasets (some from Table 1). In terms of programming technologies, we implemented our deep-learning models in Python using an in-house library that works over TensorFlow (v2.3+), HuggingFace transformers library, and integrates the W&B (Weights & Biases) platform (84) to fully track and log the developed experiments.
Post-challenge enhancements
Above, we described the ‘Annotator’ module used in the system that was evaluated within the BioCreative VII Track 2 challenge. However, posterior to the challenge, we continued studying the ‘Annotator’ behavior and theorized a set of potential improvements for this module, with some improvements being focused on efficiency and others on performance metrics. Below we present the main proposed and implemented changes along with the intuition behind them.
Masking CRF
As previously mentioned in this manuscript, we decided to follow the BIO tagging scheme to define the labels that the model could predict for each token. An interesting property of this scheme is that it encodes some structure in the sense that under no circumstance the model should predict an I (Inside) tag after an O (Outside) tag. Because, by definition, the first label of an entity should always be a B (Beginning) tag, only the B tag can appear after an O tag. This problem was naively solved in the original implementation by replacing the I tag with B whenever there was a preceding O tag. However, we argue that the model should also encode this property in its formulation, since by doing so the sequence label decoding would be more accurate, i.e. the most probable sequence |$\textbf{y*}$| should also consider this restriction. To accomplish that we manually added a mask to ft, where all the impossible transitions have a large negative weight, which according to Wei et al. (85) is equivalent to only considering the valid set of paths. In our case, we set |$\theta_t^{[\text{O,I}]}=-10\,000$|, i.e. when the previous tag was O the score of the current tag being I will be subtracted by |$10\,000$|. This weight value was empirically chosen and larger negative values did not alter the results in preliminary experiments, which indicated that |$-10\,000$| is a large enough weight for our type of sequences. Although it does not fully prevent this error from occurring, it drastically mitigates the problem leading to better model generalization as demonstrated in the ‘Results and discussion’ section.
Discarding passage splitting
In the original system, we adopted the data split format performed by the track organizers, where each article section was considered as an entire passage. However, we noticed that this caused the existence of shorter passages, for instance the title passage, and in more severe cases passages comprised only a single word, e.g. ‘Abstract’. This resulted in the creation of certain heavily padded samples, meaning that the model was not being efficiently used since given the high requirements of the transformer architecture, the amount of useful contextualized representations obtained from the transformer should be maximized. To accomplish this idea, we discarded the original passage data split and considered the whole document as an entire sequence of tokens that was fed to the model using the previously described methodology (Figure 2). As expected, this change produced much less samples per document, thus speeding up training and inference. An important note here is that by having less samples per document, there are less updates per epoch during model training, which must also be reflected in the learning rate.
Data augmentation
As a general rule of thumb in deep learning, the more data are available better the results can become. Unfortunately, human annotated data are expensive and require massive efforts to be obtained. Therefore, in deep learning it is common to see the use of data augmentation techniques, which use heuristics and randomness to increase the quantity of available data with the expectation that the new data points can help approximating the underlying real data distribution. Considering this, we experimented with two different data augmentation techniques. The first technique is inspired by Erdengasileng et al. (86), where the authors propose to randomly replace chemical entities and nonchemical tokens with random strings. The intuition to replace a chemical entity with a random string is to force the model to explore the context when predicting an entity as chemical, since the only way to correctly predict that random string as an chemical entity is with the help of the context. Additionally, to mitigate model bias on predicting random strings always as chemical entities, we also selected nonchemical tokens to be randomly replaced.
Despite the simplicity of the previously described method, it was not possible to directly implement it in our training pipeline owing to the pre-computed contextualized embeddings that we use for efficiency purposes. Therefore, we implemented a slight variation of the first data augmentation mechanism, where instead of replacing tokens with random strings we replace contextualized embeddings by random embeddings. We propose two different mechanisms for generating random embeddings: the first one is to shuffle the dimensions of the original entity sub-token embedding, which ensures that the embeddings are statistically equivalent but due to the curse of dimensionality will probably have a meaningless space position, and the second method is to generate a random embedding from a Gaussian distribution that has the same statistics of the BERT embeddings. Besides this augmentation technique that only relies on randomness to expand the datasets, we also explored a second data augmentation technique which followed a similar approach to Kim et al. (80), where the authors used the CTD table of synonyms to randomly replace some chemical entities.
Document-level agreement
Also inspired by Kim et al. (80), we experimented building a post-processing mechanism that assures entity annotation agreement throughout the whole document. For that, we followed their majority voting idea that accepts or rejects entities based on the majority voting of the model predictions in the full-text document.
Ensemble
As another rule of thumb in deep learning, using aggregated predictions of multiple models usually outperforms the predictions of a single model. This technique is known as ensemble models. Therefore, we first implemented a simple majority voting ensemble mechanism at the tagging level. More precisely, when considering multiple model predictions, we count the labels that were assigned for each token in the document, and in the end we choose the most voted label. In case of a draw, we prioritize the O and, then, B tags. However, since this technique may produce some inconsistent BIO sequences (an I tag may appear after an O tag), we propose a second majority voting technique that works at the entity level instead of the tag level, hence being aware of the entities already discovered by each of the individual models. The entity-level majority voting ensemble gathers the entities predicted by multiple models and identifies as correct predicted entities only the ones that appear in the predictions of most models.
Hyperparameter search
Finally, we employed a vast hyperparameter search to find a better configuration for our model and also to quickly understand the impact of some of the post-challenge modifications. To perform this hyperparameter search, we relied on our in-house training library that uses optuna (87) as backend. In more detail, we adopted the Tree-structured Parzen Estimator (88) sampler as the searching procedure and also used a median prunner to quickly discard bad performing trials at an early phase of training. The complete set of hyperparameters that were tuned are presented in Table 2.
NER hyperparameters using optuna on the NLM-Chem training set (‘train’ and ‘dev’ subsets). Boolean variables are True (used) or False (not used). Tests made with a maximum of 30 epochs.
| Hyperparameter . | Range of values . | Best value . |
|---|---|---|
| Trainable BERT layersa | [1, 3] | 3 |
| Sample weights | Boolean | False |
| Learning rate | [0.00001, 0.001] | 0.0003 |
| Random data augmentation | Shuffle, noise or none | Noise |
| Eb | [0.1, 0.95] | 0.66 |
| NEb | [0, 1-E] | 0.33 |
| Gaussian noise | [0.01, 0.2] | 0.15 |
| Dropout | [0.0, 0.6] | 0.3 |
| MLP dense units | [64, 1024] | 900 |
| MLP dense activation | ReLU, SeLU or Mish | Mish |
| Hyperparameter . | Range of values . | Best value . |
|---|---|---|
| Trainable BERT layersa | [1, 3] | 3 |
| Sample weights | Boolean | False |
| Learning rate | [0.00001, 0.001] | 0.0003 |
| Random data augmentation | Shuffle, noise or none | Noise |
| Eb | [0.1, 0.95] | 0.66 |
| NEb | [0, 1-E] | 0.33 |
| Gaussian noise | [0.01, 0.2] | 0.15 |
| Dropout | [0.0, 0.6] | 0.3 |
| MLP dense units | [64, 1024] | 900 |
| MLP dense activation | ReLU, SeLU or Mish | Mish |
a Last layers of the model.
b E: probability of changing an entity token representation. NE: probability of changing a non-entity token representation.
NER hyperparameters using optuna on the NLM-Chem training set (‘train’ and ‘dev’ subsets). Boolean variables are True (used) or False (not used). Tests made with a maximum of 30 epochs.
| Hyperparameter . | Range of values . | Best value . |
|---|---|---|
| Trainable BERT layersa | [1, 3] | 3 |
| Sample weights | Boolean | False |
| Learning rate | [0.00001, 0.001] | 0.0003 |
| Random data augmentation | Shuffle, noise or none | Noise |
| Eb | [0.1, 0.95] | 0.66 |
| NEb | [0, 1-E] | 0.33 |
| Gaussian noise | [0.01, 0.2] | 0.15 |
| Dropout | [0.0, 0.6] | 0.3 |
| MLP dense units | [64, 1024] | 900 |
| MLP dense activation | ReLU, SeLU or Mish | Mish |
| Hyperparameter . | Range of values . | Best value . |
|---|---|---|
| Trainable BERT layersa | [1, 3] | 3 |
| Sample weights | Boolean | False |
| Learning rate | [0.00001, 0.001] | 0.0003 |
| Random data augmentation | Shuffle, noise or none | Noise |
| Eb | [0.1, 0.95] | 0.66 |
| NEb | [0, 1-E] | 0.33 |
| Gaussian noise | [0.01, 0.2] | 0.15 |
| Dropout | [0.0, 0.6] | 0.3 |
| MLP dense units | [64, 1024] | 900 |
| MLP dense activation | ReLU, SeLU or Mish | Mish |
a Last layers of the model.
b E: probability of changing an entity token representation. NE: probability of changing a non-entity token representation.
Normalizer
After detecting chemical entities using the previous NER approach, a named entity normalization process was developed to convert entities to their corresponding MeSH codes. This normalization workflow was divided in two major components: (i) a rule-based system and (ii) a deep-learning solution based on transformers.
To supply both normalization components with curated concept-code mappings, two dictionary files were created by filtering and restructuring the 2021 MeSH and Supplementary Concept Records (SCR) files. During this filtering procedure, the MeSH file only retained concepts belonging to the ‘Drugs and Chemical’ MeSH headings subcategory, i.e. all Dxx coded categories, as these were within the scope of the present challenge.
Rule-based component
The rule-based component attempts to map entities to their corresponding MeSH codes through exact matching mechanisms. The development of this component followed an incremental workflow as described next.
For the first iteration of the rule-based system, a simple dictionary was configured that strictly used the base MeSH mappings, i.e. the DescriptorUI-DescriptorName mappings, from a subset of the MeSH 2021 filtered file. This subset comprehended mappings from the following MeSH subcategories: D01 - Inorganic Chemicals, D02 - Organic Chemicals, D03 - Heterocyclic Compounds and D04 - Polycyclic Compounds. Exact matching was then performed with this dictionary using raw text entities and lowercased entities, with the latter providing better results.
Next, to assess the impact of the mapping dictionary in system performance, the dictionary was expanded to incorporate a greater range of mappings. For that, each DescriptorUI-DescriptorName mapping from the previous dictionary was expanded, integrating related mappings from Concept Chemical Abstracts Type N1 Name (ConceptCASN1Name), which is the systematic name used in the Chemical Abstracts Chemical Substance and Formula Indexes, and also from associated entry terms, which are alternate forms or closely related terms present in the MeSH record that can be used interchangeably with the preferred term (DescriptorName) for indexing and retrieval purposes. Using the previously described procedure for augmenting the coverage of the mapping dictionary resulted in an improved performance.
As it is common to find plenty of abbreviations within biomedical literature, an abbreviation expansion step was added to the rule-based system through the integration of the Ab3P tool (89). This step was added in two different configurations, the first storing a list of previously seen abbreviations per document and a second storing the same list per corpus. When using the corpus variant, the system firstly iterates through all extracted entities and stores existing abbreviations along with their expanded form and only then it executes the exact matching pipeline. Although the document-level variant did not impact on the system performance, the inclusion of an abbreviation expansion procedure at a corpus level led to an overall improved entity-code mapping process across all training data splits.
Since there was still a significant amount of entities that the system could not map, a partial matching mechanism was added to process and map the remaining non-mapped entities. To accomplish this, the MetaMap (36)-based pyMeSHSim (55) Python package was integrated in the rule-based system. However, this partial matching mechanism was unsuccessful as (i) pyMeSHSim was very slow and thus unusable considering the large size of the test dataset and (ii) pyMeSHSim yielded numerous false positives, consequently downgrading the rule-based system performance. As a result, this partial matching mechanism was removed from the solution.
Due to the insuccess of the partial matching approach, another iteration focused on dictionary expansion was performed to further narrow down the list on unmapped entities. Since the previously selected list of MeSH subcategories (D01–D04) did not provide a complete coverage of the MeSH codes present in the gold-standard training annotations, the previous procedure of extracting mappings from the DescriptorName, ConceptCASN1Name and entry terms was repeated but considering all Dxx subcategories.
The previous procedure effectively increased MeSH code coverage, leading to increased recall without harming precision and demonstrating that the exact matching mechanism was performing as intended. Although using the whole ‘Drugs and Chemical’ subcategory allowed for a wider span of curated MeSH code mappings, it still did not manage to fully capture the MeSH codes present in the training dataset. Therefore, with the objective of addressing the existing gap between MeSH codes in the dictionary versus MeSH codes in training annotations, additional mappings for entry terms and headings related to each DescriptorName in the SCR file were added to the mapping dictionary. By exploring this novel source of information, the rule-based system obtained improved results throughout all training data splits.
At this state, the mapping dictionary already provided a large coverage of the MeSH codes present in training gold-standard annotations, with the remaining unmapped codes being mostly related with multi-word expressions that mapped to multiple MeSH codes. The remaining set of mappings was addressed with a final augmentation procedure, where complex mappings present in the gold-standard annotations (e.g. entities with multiple MeSH codes) were added to the source dictionary, ultimately improving its coverage. Finally, in the event of the rule-based mechanism failing to normalize all extracted entities, a deep-learning component was used to process the remaining unmapped entities as described next.
Deep-learning component
Following the deep-learning trend of using transformer models, we also built a complementary component relying on a dense retrieval technique to rank possible MeSH codes for each of the remaining unmapped chemical entities. Then, we apply a naive approach to decide what MeSH codes should be assigned from the ranked list. This method consists of constructing dense representations, i.e. embeddings, for each MeSH code and every predicted entity. MeSH code representations are saved in a dictionary that is stored in disk. Then, we compute the cosine similarity between the entity representations and all of the pre-computed MeSH code representations from the saved dictionary. This produces a ranked list of MeSH codes where the most similar MeSH codes for each entity take the higher positions within the ranked list. To select MeSH codes from the ranked list, a simple strategy is followed that (i) returns the MeSH code that is above a specific threshold or (ii) returns the top MeSH code if it is below the threshold but the difference between it and the second MeSH code is higher than another specific threshold. Note that this method does not ensure that all of the unmapped chemical entities are assigned a MeSH code, since it is possible to return no MeSH code if the scores do not surpass the defined threshold.
Here, we leveraged another BERT-based model to create the dense representation for the MeSH codes and for the chemical entities. More precisely, we adopted the publicly available SapBERT (90) model that was specifically fine-tuned for creating biomedical entity representation by clustering similar biomedical terms. Despite the fact that this model was not directly trained on the MeSH vocabulary, we hypothesize that the domains are closely related and thus we used it in a zero-shot fashion. To create dense representations, the detected chemical mentions and MeSH DescriptorNames were fed to SapBERT and the produced [CLS] embedding was taken as the dense representation for each chemical entity and MeSH code, respectively. These could then be compared by cosine similarity. Figure 3 presents a schematic of the deep-learning normalization model.
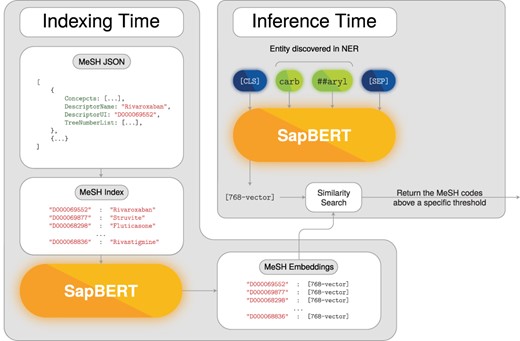
Post-challenge enhancements
Due to existing computational limitations, it was only possible to partially evaluate the deep-learning complementary component. More precisely, only two of the five submitted runs in the BioCreative VII Track 2 challenge used the complementary DL component after the rule-based component. Therefore, in a post-challenge phase we performed a deeper assessment of the DL component to evaluate its potential impact in the performance of the ‘Normalizer’ module.
Indexer
The last stage of this workflow involved selecting and indexing the MeSH codes of interest, which were previously extracted and normalized within the proposed pipeline. This task was devised in two approaches: (i) a rule-based system and (ii) a system based on TF-IDF scores.
Rule-based approach
The original rule-based approach consisted of a two-stage pipeline focused on indexing MeSH codes and was based on MeSH code location within the document, namely the title, abstract and all the captions from tables and figures. We hypothesized that these three elements of the document would contain the most relevant MeSH codes for establishing the document scope. Therefore, the initial stage gathers all of the previously extracted codes that were found in these locations.
The second stage uses this set of codes and evaluates the percentage of occurrence in the complete document. This procedure was used to narrow down the previous list of codes, which reflected positively in the precision metric on the training datasets. Rules applied in this stage had different weights for each part of the document, i.e. MeSH codes recognized in the title section required a percentage of occurrence equal or superior to 10%, MeSH codes detected in captions needed a percentage of occurrence of at least 20% and finally MeSH codes detected in the abstract required an occurrence threshold of 7%.
TF-IDF approach
This approach was inspired on the inner workings of a traditional information retrieval (IR) system, with the intuition that ultimately an IR system would be used to retrieve the documents by exploring the respective indexed MeSH codes. Therefore, we hypothesize that the indexing task can be viewed as an optimization problem of finding the limited set of MeSH codes that maximize an IR system ranking score. Furthermore, given that not every MeSH code contributes equally to the final document ranking score, we can select only the top-k codes that contribute the most as the representative MeSH codes for the document, i.e. the MeSH codes that would be indexed in order to find that a specific document.
For modeling the importance of each MeSH code we adopted the usual TF-IDF weighting schema with different SMART variations (91). More precisely, the TF-IDF schema models MeSH code importance as a function of its nonlinear frequency times its rarity. After computing the TF-IDF importance of each MeSH code per document, the next task was to select the most relevant codes. This was accomplished by using a simple-threshold-based method to select the most important MeSH codes.
Post-challenge enhancements
In a post-contest phase, the rule-based approach was modified to support the inclusion of an additional rule, responsible for evaluating MeSH codes identified in the conclusion section of the documents, when available. The augmented rule-based approach was used in an ‘unofficial’ post-contest submission, using a different set of occurrence thresholds where percentages of occurrence of 6%, 16%, 17% and 6% were selected for the title, captions, abstract and conclusion rules, respectively. Although these modifications resulted in improvements in terms of F1-score of approximately ∼5 percentage points across all data splits from the training dataset, the improved system could not surpass our official results in the test dataset, leading in fact to a small performance degradation.
However, after closing the post-contest submission phase, a more extensive analysis was performed to discover the impact of each rule. This analysis evaluated the impact of using each rule independently and of using groups of rules and was used to find the optimal occurrence thresholds for each rule. To increase the size of the development dataset, we merged the ‘train’ and ‘dev’ partitions of the training dataset, and system performance was evaluated on the ‘test’ partition of the same dataset. The best performance metrics achieved in the ‘test’ partition of the training set were obtained using a combination of the four rules and the following occurrence thresholds: 2% for the title, 22% for captions, 10% for the abstract and 10% for conclusion. The final system performance was evaluated on the actual test dataset from the NLM-Chem track.
Results and discussion
Herein, we provide a summarized view of the official submitted runs in the NLM-Chem track of the BioCreative VII challenge, present more detailed results obtained during the development of the original and improved versions of the proposed end-to-end system, discuss the impact of performed experiments and finally perform an error analysis on the resulting system.
Submitted runs
Table 3 presents the results of our official submitted runs in the different subtasks and includes additional official metrics shared by the organizers. In the normalization subtask, Runs 1, 4 and 5 used the rule-based method alone, while Runs 2 and 3 used the rule-based method followed by the deep-learning method. In the indexing subtask, Runs 1 and 5 used the rule-based approach, whereas Runs 2, 3 and 4 used the TF-IDF-based approach. The presented results demonstrated a superior performance from Run 4 in NER and normalization, meaning that it was beneficial to train in several datasets if then fine-tuned on the NLM-Chem dataset (78).
Another interesting observation was that in the normalization subtask, the rule-based method seemed to achieve high precision values while being competitive in terms of recall when compared to the median of the challenge for that subtask, giving us a comparable higher F1 measure. In terms of the last task, the rule-based approach managed to achieve competitive results, outscoring the benchmark by |$\gt4\%$| points. On the other hand, TF-IDF did not manage to beat the benchmark, showing an overall poor performance, which may disprove the main hypothesis behind the idea or that the naive approach is too simple to model this problem.
For a more detailed technical description of the submitted runs for each subtask, please refer to our proceedings paper (78).
Official obtained results on the final NLM-Chem test set (evaluation dataset). All the results presented use the strict evaluation method. Our top score results are highlighted in bold.
| . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Chemical mention recognition | |||
| Run 1 | 0.8354 | 0.8429 | 0.8392 |
| Run 2 | 0.8421 | 0.8350 | 0.8386 |
| Run 3 | 0.8505 | 0.7662 | 0.8062 |
| Run 4 | 0.8394 | 0.8515 | 0.8454 |
| Run 5 | 0.8372 | 0.7416 | 0.7865 |
| Median | 0.8476 | 0.8136 | 0.8373 |
| Benchmark | 0.8440 | 0.7877 | 0.8149 |
| Chemical normalization to MeSH IDs | |||
| Run 1 | 0.8582 | 0.7641 | 0.8084 |
| Run 2 | 0.8221 | 0.7898 | 0.8056 |
| Run 3 | 0.8124 | 0.7760 | 0.7938 |
| Run 4 | 0.8621 | 0.7702 | 0.8136 |
| Run 5 | 0.8310 | 0.7411 | 0.7835 |
| Median | 0.7120 | 0.7760 | 0.7749 |
| Benchmark | 0.8151 | 0.7644 | 0.7889 |
| Chemical indexing | |||
| Run 1 | 0.5351 | 0.4133 | 0.4664 |
| Run 2 | 0.4882 | 0.3284 | 0.3927 |
| Run 3 | 0.4910 | 0.3236 | 0.3901 |
| Run 4 | 0.5173 | 0.3236 | 0.3981 |
| Run 5 | 0.5308 | 0.3812 | 0.4437 |
| Median | 0.5173 | 0.3284 | 0.3981 |
| Benchmark | 0.3134 | 0.6101 | 0.4141 |
| . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Chemical mention recognition | |||
| Run 1 | 0.8354 | 0.8429 | 0.8392 |
| Run 2 | 0.8421 | 0.8350 | 0.8386 |
| Run 3 | 0.8505 | 0.7662 | 0.8062 |
| Run 4 | 0.8394 | 0.8515 | 0.8454 |
| Run 5 | 0.8372 | 0.7416 | 0.7865 |
| Median | 0.8476 | 0.8136 | 0.8373 |
| Benchmark | 0.8440 | 0.7877 | 0.8149 |
| Chemical normalization to MeSH IDs | |||
| Run 1 | 0.8582 | 0.7641 | 0.8084 |
| Run 2 | 0.8221 | 0.7898 | 0.8056 |
| Run 3 | 0.8124 | 0.7760 | 0.7938 |
| Run 4 | 0.8621 | 0.7702 | 0.8136 |
| Run 5 | 0.8310 | 0.7411 | 0.7835 |
| Median | 0.7120 | 0.7760 | 0.7749 |
| Benchmark | 0.8151 | 0.7644 | 0.7889 |
| Chemical indexing | |||
| Run 1 | 0.5351 | 0.4133 | 0.4664 |
| Run 2 | 0.4882 | 0.3284 | 0.3927 |
| Run 3 | 0.4910 | 0.3236 | 0.3901 |
| Run 4 | 0.5173 | 0.3236 | 0.3981 |
| Run 5 | 0.5308 | 0.3812 | 0.4437 |
| Median | 0.5173 | 0.3284 | 0.3981 |
| Benchmark | 0.3134 | 0.6101 | 0.4141 |
Official obtained results on the final NLM-Chem test set (evaluation dataset). All the results presented use the strict evaluation method. Our top score results are highlighted in bold.
| . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Chemical mention recognition | |||
| Run 1 | 0.8354 | 0.8429 | 0.8392 |
| Run 2 | 0.8421 | 0.8350 | 0.8386 |
| Run 3 | 0.8505 | 0.7662 | 0.8062 |
| Run 4 | 0.8394 | 0.8515 | 0.8454 |
| Run 5 | 0.8372 | 0.7416 | 0.7865 |
| Median | 0.8476 | 0.8136 | 0.8373 |
| Benchmark | 0.8440 | 0.7877 | 0.8149 |
| Chemical normalization to MeSH IDs | |||
| Run 1 | 0.8582 | 0.7641 | 0.8084 |
| Run 2 | 0.8221 | 0.7898 | 0.8056 |
| Run 3 | 0.8124 | 0.7760 | 0.7938 |
| Run 4 | 0.8621 | 0.7702 | 0.8136 |
| Run 5 | 0.8310 | 0.7411 | 0.7835 |
| Median | 0.7120 | 0.7760 | 0.7749 |
| Benchmark | 0.8151 | 0.7644 | 0.7889 |
| Chemical indexing | |||
| Run 1 | 0.5351 | 0.4133 | 0.4664 |
| Run 2 | 0.4882 | 0.3284 | 0.3927 |
| Run 3 | 0.4910 | 0.3236 | 0.3901 |
| Run 4 | 0.5173 | 0.3236 | 0.3981 |
| Run 5 | 0.5308 | 0.3812 | 0.4437 |
| Median | 0.5173 | 0.3284 | 0.3981 |
| Benchmark | 0.3134 | 0.6101 | 0.4141 |
| . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Chemical mention recognition | |||
| Run 1 | 0.8354 | 0.8429 | 0.8392 |
| Run 2 | 0.8421 | 0.8350 | 0.8386 |
| Run 3 | 0.8505 | 0.7662 | 0.8062 |
| Run 4 | 0.8394 | 0.8515 | 0.8454 |
| Run 5 | 0.8372 | 0.7416 | 0.7865 |
| Median | 0.8476 | 0.8136 | 0.8373 |
| Benchmark | 0.8440 | 0.7877 | 0.8149 |
| Chemical normalization to MeSH IDs | |||
| Run 1 | 0.8582 | 0.7641 | 0.8084 |
| Run 2 | 0.8221 | 0.7898 | 0.8056 |
| Run 3 | 0.8124 | 0.7760 | 0.7938 |
| Run 4 | 0.8621 | 0.7702 | 0.8136 |
| Run 5 | 0.8310 | 0.7411 | 0.7835 |
| Median | 0.7120 | 0.7760 | 0.7749 |
| Benchmark | 0.8151 | 0.7644 | 0.7889 |
| Chemical indexing | |||
| Run 1 | 0.5351 | 0.4133 | 0.4664 |
| Run 2 | 0.4882 | 0.3284 | 0.3927 |
| Run 3 | 0.4910 | 0.3236 | 0.3901 |
| Run 4 | 0.5173 | 0.3236 | 0.3981 |
| Run 5 | 0.5308 | 0.3812 | 0.4437 |
| Median | 0.5173 | 0.3284 | 0.3981 |
| Benchmark | 0.3134 | 0.6101 | 0.4141 |
Chemical recognition
As previously mentioned, the ‘Annotator’ is responsible for addressing the chemical recognition problem, acting as the first stage of our system pipeline. As a consequence, any error produced by the ‘Annotator’ is forwarded to downstream modules, lowering the system’s overall performance. Therefore, a significant part of this work was invested on the evaluation of several configurations with the objective of building an efficient yet performing NER model. Here, we firstly present a summary of the experiments conducted during NER model development for the BioCreative VII Track 2 challenge, followed by the new post-challenge modifications and their impact on the official test set.
Table 4 presents a brief summary of the incremental changes made to the NER model that contributed to the most substantial improvement of the validation metrics recorded over the NLM-Chem ‘test’ subset of the training set. In short, we began with the plain PubMedBERT contextualized embeddings (PubMedBERT was not trained) and trained a linear classifier on the NLM-Chem ‘train’ and ‘dev’ subsets. This approach provided us the baseline F1-score of 0.7140. Next, we tried to increase the complexity of our classifier by replacing the linear classifier by a MLP or CNN. In the end, we found that the MLP classifier led to better performances, achieving an F1-score of 0.7539. Up to this point our NER model was using the independence assumption to estimate tags (Equation 1). However, as aforementioned, that assumption does not hold under this problem. Thus, following Equation 3, a CRF classifier was added after the MLP, leading to a performance gain of 4.8% points in F1-score, bringing our best result to 0.8020. The final considerable improvement was achieved once we started training the last layer of PubMedBERT. Additionally, since each transformer block contains millions of parameters, we built a larger training corpus to prevent the model from overfitting on the NLM-Chem dataset, which also provided more previously unseen chemical entities for the model to explore. With more detail, the full corpus contained datasets from well-known chemical datasets for NER, and the complete list can be consulted in our proceedings paper (78). At last, we pre-trained our model on the previously collected datasets with the exception of NLM-Chem, which was used for fine-tuning. This model achieved a new best result of 0.8584 of F1-score, and it was the model used in our best submission to the BioCreative VII Track 2 challenge (Run 4).
NER ablation study conducted during the BioCreative VII track 2 challenge in the NLM-Chem training set (‘test’ subset).
| . | F1-score . |
|---|---|
| PubMedBERT (frozen) + Linear classifier | 0.7140 |
| + MLP classifier | 0.7539 |
| + CRF classifier | 0.8020 |
| + PubMedBERT (unfrozen last layer) + pre-train and fine-tunea | 0.8584 |
| . | F1-score . |
|---|---|
| PubMedBERT (frozen) + Linear classifier | 0.7140 |
| + MLP classifier | 0.7539 |
| + CRF classifier | 0.8020 |
| + PubMedBERT (unfrozen last layer) + pre-train and fine-tunea | 0.8584 |
a Pre-training corresponds in firstly training the model in entity recognition using several chemical NER datasets described in our challenge paper (78). Then, the fine-tune step corresponds in further training the model for 5 epochs on the NLM-Chem dataset (‘train’ and ‘dev’ subsets).
NER ablation study conducted during the BioCreative VII track 2 challenge in the NLM-Chem training set (‘test’ subset).
| . | F1-score . |
|---|---|
| PubMedBERT (frozen) + Linear classifier | 0.7140 |
| + MLP classifier | 0.7539 |
| + CRF classifier | 0.8020 |
| + PubMedBERT (unfrozen last layer) + pre-train and fine-tunea | 0.8584 |
| . | F1-score . |
|---|---|
| PubMedBERT (frozen) + Linear classifier | 0.7140 |
| + MLP classifier | 0.7539 |
| + CRF classifier | 0.8020 |
| + PubMedBERT (unfrozen last layer) + pre-train and fine-tunea | 0.8584 |
a Pre-training corresponds in firstly training the model in entity recognition using several chemical NER datasets described in our challenge paper (78). Then, the fine-tune step corresponds in further training the model for 5 epochs on the NLM-Chem dataset (‘train’ and ‘dev’ subsets).
Post-challenge enhancements analysis
With the objective of further improving the model in a post-challenge stage, a set of changes was defined that could lead to gains in terms of efficiency and performance metrics.
Firstly, we proposed the addition of a mask to the CRF classifier transition matrix to penalize impossible transitions, for instance the appearance of an I tag after an O tag. To measure the impact of this technique, we performed an experiment where the same model was trained under the same conditions except for the presence or absence of the aforementioned CRF mask. Figure 4 presents the results of this experiment, where we evaluated the F1-score on the NLM-Chem test subset and counted the number of times that the model wrongfully predicted an I tag after an O tag. As observable, using a CRF mask not only led to consistently higher F1-score, but more interestingly it drastically reduced the number of times that the model predicted an I tag after an O tag, effectively demonstrating the success of the mask in mitigating this error. This result also shows that by directly learning how to avoid this mistake the model was able to better generalize, as suggested by the consistently higher F1-score. It is also important to mention that in both models our decoding mechanism automatically converted the incorrect I tags (which appear after an O tag) to B tags. However, this mechanism seems to be more biased toward the model that did not use CRF mask, since the model is not penalized in terms of F1-score in situations where it wrongly predicts an I tag after an O tag instead of a B tag. Nevertheless, even with the presence of such bias the model with CRF mask managed to consistently outscore the model without it.
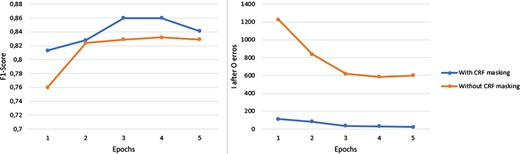
Performance comparison between the use or absence of a CRF mask. The measurements were taken during the five initial training epochs of the exactly same model and training configuration. The only variable that changed was the CRF mask.
Next, Table 5 compares the adopted strategy for converting full-text articles into individual samples that the model can process, which was named as data generator. The first row presents the number of samples produced by the data generator used during the official challenge, whereas the second row refers to the post-challenge version that discards the passage splitting step. It is noticeable that the previous data generator produced over 2.5 times more samples than the new version. This difference means that our model can now process the same amount of data, but 2.5 times faster. Furthermore, it is also important to consider that each training epoch now has 2.5 times less samples, corresponding to less training updates and meaning that certain adjustments have to be performed to train the model with this new data generator. Finally, we did not notice any significant improvement in F1-score from using the new data generator instead of its old counterpart.
Comparison between the data generator used during the BioCreative NLM-Chem challenge and the post-challenge improvement that proposes to discard the document passage split and directly use full-text. This shows the number of samples produced by each method in each subset of the NLM-Chem training set.
| . | Number of samples for each NLM-Chem subset . | |||
|---|---|---|---|---|
| . | Train . | Dev . | Test . | Total . |
| Previous data preparation | 6299 | 1472 | 3858 | 11 629 |
| + Discard passage split | 2351 | 572 | 1338 | 4264 |
| . | Number of samples for each NLM-Chem subset . | |||
|---|---|---|---|---|
| . | Train . | Dev . | Test . | Total . |
| Previous data preparation | 6299 | 1472 | 3858 | 11 629 |
| + Discard passage split | 2351 | 572 | 1338 | 4264 |
Comparison between the data generator used during the BioCreative NLM-Chem challenge and the post-challenge improvement that proposes to discard the document passage split and directly use full-text. This shows the number of samples produced by each method in each subset of the NLM-Chem training set.
| . | Number of samples for each NLM-Chem subset . | |||
|---|---|---|---|---|
| . | Train . | Dev . | Test . | Total . |
| Previous data preparation | 6299 | 1472 | 3858 | 11 629 |
| + Discard passage split | 2351 | 572 | 1338 | 4264 |
| . | Number of samples for each NLM-Chem subset . | |||
|---|---|---|---|---|
| . | Train . | Dev . | Test . | Total . |
| Previous data preparation | 6299 | 1472 | 3858 | 11 629 |
| + Discard passage split | 2351 | 572 | 1338 | 4264 |
In another post-challenge enhancement we conducted a vast hyperparameter search comprising a total of 424 trials to find a good hyperparameter set and also to assess the impact of different hyperparameters. Table 2 shows the different hyperparameters considered during the search along with their range of values and the final chosen value. This search was performed using the CRF mask and the new data generator, as both aspects had already demonstrated their positive impact in system performance. Upon inspection of logged values from executed runs we concluded the following:
Increasing the number of trainable layers from PubMedBERT provided consistent gains in terms of F1-score. More precisely, by training only the last layer the maximum achieved score was no higher than 0.86 on the NLM-Chem ‘test’ subset, while by training the last three layers we managed to consistently achieve F1-scores above 0.87. Due to computational limitations we only managed to perform training until the antepenultimate layer of the PubMedBERT model.
Random data augmentation seemed to be beneficial, although without a clear difference between the two methods (shuffle and noise).
Adding a Gaussian noise layer after the contextualized embedding resulted in consistent improvements.
The non-monotonic Mish activation function outperformed any other activation function during hyperparameter search experiments.
Following the best model configurations and intuition gathered during the hyperparameter search procedure, further experiments were performed to assess whether it was beneficial to use more chemical datasets as training data (Table 6), similarly to what was done during the BioCreative challenge. Additionally, we also aimed to see the gains that the previously described majority voting ensemble methods could achieve. Therefore, three different pre-training scenarios were considered using: (1) no additional data; (2) CCD datasets, which is short for CDR, CHEMDNER and DrugProt, and (3) CCD and NLM-Chem-Syn. After all three pre-training scenarios, models were fine-tuned on the (i) ‘train’ and ‘dev’ subsets or (ii) ‘train’, ‘dev’ and ‘test’ subsets of the NLM-Chem. Note that in experiments using the test subset it is not sensible to measure system performance on the same subset, hence the missing values in Table 6. Combining the three pre-training methods with the two fine-tuning methods resulted in six unique experiments, and for each experiment we trained five models with different random seeds. Then, we reported the average and standard deviation of the performance of the five models, and additionally we used the same five models to compute the tag-level majority voting (T) and the entity-level majority voting (E).
NER results, using strict evaluation, obtained from extensive post-challenge experiments. All the models were trained for 20 epochs. P: pre-training. T: indication if the NLM-Chem ‘test’ subset (training set) was used in the fine-tuning stage, and the NLM-Chem-Syn ‘test’ subset was used in the pre-training. E: indication of the ensemble method used—tag-level (T) or entity-level (E) majority voting. CCD: CDR, CHEMDNER and DrugProt. Syn: synthetic NLM-Chem dataset. Standard deviation is presented in parentheses. The highest F1-score results are highlighted in bold.
| . | . | . | Evaluation on the . | . | Evaluation on the . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | NLM-Chem training set (‘test’ subset) . | . | final NLM-Chem test set . | ||||
| Pa . | Tb . | Ec . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| – | N | – | 0.8497 (0.0047) | 0.8807 (0.0044) | 0.8649 (0.0032) | 0.8468 (0.0044) | 0.8673 (0.0015) | 0.8569 (0.0017) | |
| – | N | T | 0.8530 | 0.8861 | 0.8692 | 0.8508 | 0.8718 | 0.8612 | |
| – | N | E | 0.8573 | 0.8846 | 0.8707 | 0.8573 | 0.8699 | 0.8636 | |
| CCD | N | – | 0.8590 (0.0035) | 0.8837 (0.0028) | 0.8712 (0.0017) | 0.8586 (0.0052) | 0.8667 (0.0022) | 0.8626 (0.0031) | |
| CCD | N | T | 0.8658 | 0.8931 | 0.8792 | 0.8643 | 0.8726 | 0.8685 | |
| CCD | N | E | 0.8685 | 0.8906 | 0.8794 | 0.8721 | 0.8714 | 0.8717 | |
| CCD, Syn | N | – | 0.8579 (0.0021) | 0.8818 (0.0016) | 0.8696 (0.0018) | 0.8569 (0.0035) | 0.8589 (0.0032) | 0.8579 (0.0013) | |
| CCD, Syn | N | T | 0.8625 | 0.8890 | 0.8755 | 0.8604 | 0.8631 | 0.8617 | |
| CCD, Syn | N | E | 0.8675 | 0.8873 | 0.8773 | 0.8668 | 0.8620 | 0.8644 | |
| – | Y | – | – | – | – | 0.8565 (0.0021) | 0.8634 (0.0010) | 0.8600 (0.0012) | |
| – | Y | T | – | – | – | 0.8604 | 0.8672 | 0.8638 | |
| – | Y | E | – | – | – | 0.8655 | 0.8659 | 0.8657 | |
| CCD | Y | – | – | – | – | 0.8669 (0.0023) | 0.8648 (0.0028) | 0.8659 (0.0022) | |
| CCD | Y | T | – | – | – | 0.8713 | 0.8704 | 0.8708 | |
| CCD | Y | E | – | – | – | 0.8775 | 0.8688 | 0.8731 | |
| CCD, Syn | Y | – | – | – | – | 0.8627 (0.0034) | 0.8564 (0.0047) | 0.8594 (0.0019) | |
| CCD, Syn | Y | T | – | – | – | 0.8663 | 0.8612 | 0.8637 | |
| CCD, Syn | Y | E | – | – | – | 0.8715 | 0.8601 | 0.8658 | |
| . | . | . | Evaluation on the . | . | Evaluation on the . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | NLM-Chem training set (‘test’ subset) . | . | final NLM-Chem test set . | ||||
| Pa . | Tb . | Ec . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| – | N | – | 0.8497 (0.0047) | 0.8807 (0.0044) | 0.8649 (0.0032) | 0.8468 (0.0044) | 0.8673 (0.0015) | 0.8569 (0.0017) | |
| – | N | T | 0.8530 | 0.8861 | 0.8692 | 0.8508 | 0.8718 | 0.8612 | |
| – | N | E | 0.8573 | 0.8846 | 0.8707 | 0.8573 | 0.8699 | 0.8636 | |
| CCD | N | – | 0.8590 (0.0035) | 0.8837 (0.0028) | 0.8712 (0.0017) | 0.8586 (0.0052) | 0.8667 (0.0022) | 0.8626 (0.0031) | |
| CCD | N | T | 0.8658 | 0.8931 | 0.8792 | 0.8643 | 0.8726 | 0.8685 | |
| CCD | N | E | 0.8685 | 0.8906 | 0.8794 | 0.8721 | 0.8714 | 0.8717 | |
| CCD, Syn | N | – | 0.8579 (0.0021) | 0.8818 (0.0016) | 0.8696 (0.0018) | 0.8569 (0.0035) | 0.8589 (0.0032) | 0.8579 (0.0013) | |
| CCD, Syn | N | T | 0.8625 | 0.8890 | 0.8755 | 0.8604 | 0.8631 | 0.8617 | |
| CCD, Syn | N | E | 0.8675 | 0.8873 | 0.8773 | 0.8668 | 0.8620 | 0.8644 | |
| – | Y | – | – | – | – | 0.8565 (0.0021) | 0.8634 (0.0010) | 0.8600 (0.0012) | |
| – | Y | T | – | – | – | 0.8604 | 0.8672 | 0.8638 | |
| – | Y | E | – | – | – | 0.8655 | 0.8659 | 0.8657 | |
| CCD | Y | – | – | – | – | 0.8669 (0.0023) | 0.8648 (0.0028) | 0.8659 (0.0022) | |
| CCD | Y | T | – | – | – | 0.8713 | 0.8704 | 0.8708 | |
| CCD | Y | E | – | – | – | 0.8775 | 0.8688 | 0.8731 | |
| CCD, Syn | Y | – | – | – | – | 0.8627 (0.0034) | 0.8564 (0.0047) | 0.8594 (0.0019) | |
| CCD, Syn | Y | T | – | – | – | 0.8663 | 0.8612 | 0.8637 | |
| CCD, Syn | Y | E | – | – | – | 0.8715 | 0.8601 | 0.8658 | |
a Pre-training corresponds to the first training-pass of the deep-learning model (20 epochs).
b ‘N’ (No) means that the NLM-Chem ‘test’ subset (training set) was not used in the last training pass, and the NLM-Chem-Syn ‘test’ subset was not used during pre-training. ‘Y’ (Yes) means that the ‘test’ subset of the NLM-Chem and NLM-Chem-Syn datasets were used in the last training-pass and pre-training, respectively.
c An ensemble of five different models trained with different random seeds. Majority voting was applied at the T or E. The rows in which ensemble was not used (-) present the average results of the same five models (standard deviation is shown in parentheses).
NER results, using strict evaluation, obtained from extensive post-challenge experiments. All the models were trained for 20 epochs. P: pre-training. T: indication if the NLM-Chem ‘test’ subset (training set) was used in the fine-tuning stage, and the NLM-Chem-Syn ‘test’ subset was used in the pre-training. E: indication of the ensemble method used—tag-level (T) or entity-level (E) majority voting. CCD: CDR, CHEMDNER and DrugProt. Syn: synthetic NLM-Chem dataset. Standard deviation is presented in parentheses. The highest F1-score results are highlighted in bold.
| . | . | . | Evaluation on the . | . | Evaluation on the . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | NLM-Chem training set (‘test’ subset) . | . | final NLM-Chem test set . | ||||
| Pa . | Tb . | Ec . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| – | N | – | 0.8497 (0.0047) | 0.8807 (0.0044) | 0.8649 (0.0032) | 0.8468 (0.0044) | 0.8673 (0.0015) | 0.8569 (0.0017) | |
| – | N | T | 0.8530 | 0.8861 | 0.8692 | 0.8508 | 0.8718 | 0.8612 | |
| – | N | E | 0.8573 | 0.8846 | 0.8707 | 0.8573 | 0.8699 | 0.8636 | |
| CCD | N | – | 0.8590 (0.0035) | 0.8837 (0.0028) | 0.8712 (0.0017) | 0.8586 (0.0052) | 0.8667 (0.0022) | 0.8626 (0.0031) | |
| CCD | N | T | 0.8658 | 0.8931 | 0.8792 | 0.8643 | 0.8726 | 0.8685 | |
| CCD | N | E | 0.8685 | 0.8906 | 0.8794 | 0.8721 | 0.8714 | 0.8717 | |
| CCD, Syn | N | – | 0.8579 (0.0021) | 0.8818 (0.0016) | 0.8696 (0.0018) | 0.8569 (0.0035) | 0.8589 (0.0032) | 0.8579 (0.0013) | |
| CCD, Syn | N | T | 0.8625 | 0.8890 | 0.8755 | 0.8604 | 0.8631 | 0.8617 | |
| CCD, Syn | N | E | 0.8675 | 0.8873 | 0.8773 | 0.8668 | 0.8620 | 0.8644 | |
| – | Y | – | – | – | – | 0.8565 (0.0021) | 0.8634 (0.0010) | 0.8600 (0.0012) | |
| – | Y | T | – | – | – | 0.8604 | 0.8672 | 0.8638 | |
| – | Y | E | – | – | – | 0.8655 | 0.8659 | 0.8657 | |
| CCD | Y | – | – | – | – | 0.8669 (0.0023) | 0.8648 (0.0028) | 0.8659 (0.0022) | |
| CCD | Y | T | – | – | – | 0.8713 | 0.8704 | 0.8708 | |
| CCD | Y | E | – | – | – | 0.8775 | 0.8688 | 0.8731 | |
| CCD, Syn | Y | – | – | – | – | 0.8627 (0.0034) | 0.8564 (0.0047) | 0.8594 (0.0019) | |
| CCD, Syn | Y | T | – | – | – | 0.8663 | 0.8612 | 0.8637 | |
| CCD, Syn | Y | E | – | – | – | 0.8715 | 0.8601 | 0.8658 | |
| . | . | . | Evaluation on the . | . | Evaluation on the . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | NLM-Chem training set (‘test’ subset) . | . | final NLM-Chem test set . | ||||
| Pa . | Tb . | Ec . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| – | N | – | 0.8497 (0.0047) | 0.8807 (0.0044) | 0.8649 (0.0032) | 0.8468 (0.0044) | 0.8673 (0.0015) | 0.8569 (0.0017) | |
| – | N | T | 0.8530 | 0.8861 | 0.8692 | 0.8508 | 0.8718 | 0.8612 | |
| – | N | E | 0.8573 | 0.8846 | 0.8707 | 0.8573 | 0.8699 | 0.8636 | |
| CCD | N | – | 0.8590 (0.0035) | 0.8837 (0.0028) | 0.8712 (0.0017) | 0.8586 (0.0052) | 0.8667 (0.0022) | 0.8626 (0.0031) | |
| CCD | N | T | 0.8658 | 0.8931 | 0.8792 | 0.8643 | 0.8726 | 0.8685 | |
| CCD | N | E | 0.8685 | 0.8906 | 0.8794 | 0.8721 | 0.8714 | 0.8717 | |
| CCD, Syn | N | – | 0.8579 (0.0021) | 0.8818 (0.0016) | 0.8696 (0.0018) | 0.8569 (0.0035) | 0.8589 (0.0032) | 0.8579 (0.0013) | |
| CCD, Syn | N | T | 0.8625 | 0.8890 | 0.8755 | 0.8604 | 0.8631 | 0.8617 | |
| CCD, Syn | N | E | 0.8675 | 0.8873 | 0.8773 | 0.8668 | 0.8620 | 0.8644 | |
| – | Y | – | – | – | – | 0.8565 (0.0021) | 0.8634 (0.0010) | 0.8600 (0.0012) | |
| – | Y | T | – | – | – | 0.8604 | 0.8672 | 0.8638 | |
| – | Y | E | – | – | – | 0.8655 | 0.8659 | 0.8657 | |
| CCD | Y | – | – | – | – | 0.8669 (0.0023) | 0.8648 (0.0028) | 0.8659 (0.0022) | |
| CCD | Y | T | – | – | – | 0.8713 | 0.8704 | 0.8708 | |
| CCD | Y | E | – | – | – | 0.8775 | 0.8688 | 0.8731 | |
| CCD, Syn | Y | – | – | – | – | 0.8627 (0.0034) | 0.8564 (0.0047) | 0.8594 (0.0019) | |
| CCD, Syn | Y | T | – | – | – | 0.8663 | 0.8612 | 0.8637 | |
| CCD, Syn | Y | E | – | – | – | 0.8715 | 0.8601 | 0.8658 | |
a Pre-training corresponds to the first training-pass of the deep-learning model (20 epochs).
b ‘N’ (No) means that the NLM-Chem ‘test’ subset (training set) was not used in the last training pass, and the NLM-Chem-Syn ‘test’ subset was not used during pre-training. ‘Y’ (Yes) means that the ‘test’ subset of the NLM-Chem and NLM-Chem-Syn datasets were used in the last training-pass and pre-training, respectively.
c An ensemble of five different models trained with different random seeds. Majority voting was applied at the T or E. The rows in which ensemble was not used (-) present the average results of the same five models (standard deviation is shown in parentheses).
When inspecting Table 6, the first important observation is that both majority voting ensemble methods seem to perform surprisingly well, since in all of the experiments it managed to outperform the mean plus the standard deviation of the respective five models. Furthermore, the entity-level majority voting method also slightly outperformed the tagging-level majority voting, which supports the idea that the tagging level may produce inconsistent and invalid BIO sequences causing ruptured entities. Next, when looking at the results reported on the NLM-Chem ‘test’ subset it seems that using pre-training data is beneficial for system performance, with CCD being preferable over CCD and NLM-Chem-Syn, thus showing that using CTD synonym augmentation was not beneficial. Another interesting conclusion is that using the ‘test’ subset as training data led to a marginal increase from 0.8569 to 0.8600. Thus, at this point performance gains become questionable since adding 50% more training documents only resulted in a 0.0031 increase in F1-score. Finally, our best model achieved a F1-score of 0.8731 in the final NLM-Chem test set, and was based on an entity-level ensemble of five models that were pre-trained on the CCD datasets and then fine-tuned on the NLM-Chem ‘train’, ‘dev’ and ‘test’ subsets.
As a final remark, the document-level agreement technique did not manage to achieve satisfactory results during all of our experiments. It is our understanding that for this technique to work, one should also consider the annotation guidelines to better understand and further improve the mechanism.
Chemical normalization
The process of normalizing detected chemical entities was heavily reliant on a rule-based system, which performed exact matching supported by a custom dictionary that mapped textual entities into MeSH codes. As previously mentioned in the Methodology section, this mapping dictionary was created through an iterative procedure where the dictionary was progressively adjusted and improved. Table 7 summarizes system performances obtained on the training dataset during this iterative development process.
Results obtained during the iterative development process of the rule-based normalization approach in the training dataset. Dictionaries used in exact matching contained mappings for DescriptorName, ConceptCASN1Name and Entry Terms. Best results are highlighted in bold.
| Config. . | Matching . | Lowercase . | Abbreviaton expansion level . | Valid MeSH tree subcategories . | SCR . | Precision . | Recall . | F1-Score . |
|---|---|---|---|---|---|---|---|---|
| 1 | Exact | No | None | D01, D02, D03, D04 | No | 0.9389 | 0.3092 | 0.4562 |
| 2 | Exact | Yes | None | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 3 | Exact | Yes | Document | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 4 | Exact | Yes | Corpus | D01, D02, D03, D04 | No | 0.9316 | 0.3819 | 0.5417 |
| 5 | Exact + Partiala | Yes | Corpus | D01, D02, D03, D04 | No | 0.8426 | 0.3887 | 0.5320 |
| 6 | Exact | Yes | Corpus | All Dxx | No | 0.9361 | 0.5160 | 0.6653 |
| 7 | Exact | Yes | Corpus | All Dxx | Yes | 0.9439 | 0.6594 | 0.7764 |
| 8 | Exactb | Yes | Corpus | All Dxx | Yes | 0.9375 | 0.7957 | 0.8608 |
| Config. . | Matching . | Lowercase . | Abbreviaton expansion level . | Valid MeSH tree subcategories . | SCR . | Precision . | Recall . | F1-Score . |
|---|---|---|---|---|---|---|---|---|
| 1 | Exact | No | None | D01, D02, D03, D04 | No | 0.9389 | 0.3092 | 0.4562 |
| 2 | Exact | Yes | None | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 3 | Exact | Yes | Document | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 4 | Exact | Yes | Corpus | D01, D02, D03, D04 | No | 0.9316 | 0.3819 | 0.5417 |
| 5 | Exact + Partiala | Yes | Corpus | D01, D02, D03, D04 | No | 0.8426 | 0.3887 | 0.5320 |
| 6 | Exact | Yes | Corpus | All Dxx | No | 0.9361 | 0.5160 | 0.6653 |
| 7 | Exact | Yes | Corpus | All Dxx | Yes | 0.9439 | 0.6594 | 0.7764 |
| 8 | Exactb | Yes | Corpus | All Dxx | Yes | 0.9375 | 0.7957 | 0.8608 |
a Partial matching performed using pyMeSHSim, which integrates MetaMap.
b Mapping dictionary augmented with complex gold-standard mappings from the train and development partitions of the training dataset.
Results obtained during the iterative development process of the rule-based normalization approach in the training dataset. Dictionaries used in exact matching contained mappings for DescriptorName, ConceptCASN1Name and Entry Terms. Best results are highlighted in bold.
| Config. . | Matching . | Lowercase . | Abbreviaton expansion level . | Valid MeSH tree subcategories . | SCR . | Precision . | Recall . | F1-Score . |
|---|---|---|---|---|---|---|---|---|
| 1 | Exact | No | None | D01, D02, D03, D04 | No | 0.9389 | 0.3092 | 0.4562 |
| 2 | Exact | Yes | None | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 3 | Exact | Yes | Document | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 4 | Exact | Yes | Corpus | D01, D02, D03, D04 | No | 0.9316 | 0.3819 | 0.5417 |
| 5 | Exact + Partiala | Yes | Corpus | D01, D02, D03, D04 | No | 0.8426 | 0.3887 | 0.5320 |
| 6 | Exact | Yes | Corpus | All Dxx | No | 0.9361 | 0.5160 | 0.6653 |
| 7 | Exact | Yes | Corpus | All Dxx | Yes | 0.9439 | 0.6594 | 0.7764 |
| 8 | Exactb | Yes | Corpus | All Dxx | Yes | 0.9375 | 0.7957 | 0.8608 |
| Config. . | Matching . | Lowercase . | Abbreviaton expansion level . | Valid MeSH tree subcategories . | SCR . | Precision . | Recall . | F1-Score . |
|---|---|---|---|---|---|---|---|---|
| 1 | Exact | No | None | D01, D02, D03, D04 | No | 0.9389 | 0.3092 | 0.4562 |
| 2 | Exact | Yes | None | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 3 | Exact | Yes | Document | D01, D02, D03, D04 | No | 0.9424 | 0.3707 | 0.5321 |
| 4 | Exact | Yes | Corpus | D01, D02, D03, D04 | No | 0.9316 | 0.3819 | 0.5417 |
| 5 | Exact + Partiala | Yes | Corpus | D01, D02, D03, D04 | No | 0.8426 | 0.3887 | 0.5320 |
| 6 | Exact | Yes | Corpus | All Dxx | No | 0.9361 | 0.5160 | 0.6653 |
| 7 | Exact | Yes | Corpus | All Dxx | Yes | 0.9439 | 0.6594 | 0.7764 |
| 8 | Exactb | Yes | Corpus | All Dxx | Yes | 0.9375 | 0.7957 | 0.8608 |
a Partial matching performed using pyMeSHSim, which integrates MetaMap.
b Mapping dictionary augmented with complex gold-standard mappings from the train and development partitions of the training dataset.
All of the results presented in Table 7 involved the use of dictionaries with MeSH mappings for the DescriptorName, ConceptCASN1Name and entry terms. Although the first iteration actually involved a simpler dictionary containing only DescriptorName mappings (from D01–D04 MeSH subcategories), the corresponding system performance was marginal with only ∼1% of the training entities being mapped, hence not being reported in Table 7.
The first system presented in Table 7 attained a F1-score of 0.4562, showing the expected high precision of an exact matching system (0.9389) but having a low recall (0.3092) that penalized the F1-score. The second row presents a similar configuration where the matching procedure was adjusted to use lowercased instead of raw text entities, which led to improvements across all metrics but still attained a reduced recall (0.3707). In the third and fourth configurations, the system from Configuration 2 was augmented with an abbreviation expansion mechanism that can work at two different levels: document or corpus level. While the document-level configuration provided no gains in terms of performance, the corpus-level variant led to a slight improvement in recall and F1-score at the cost of a decrease in precision.
The fifth configuration presents a system configuration where partial matching was explored, through the integration of pyMeSHSim, with the objective of addressing the low recall verified in the previous system configurations. The inclusion of partial matching did not lead to the expected performance improvement, resulting in a 0.007% point gain in recall and a reduction in precision close to 0.1% points, demonstrating a significant increase in false positives.
The sixth system configuration built on top of the fourth configuration, removing the partial matching component and expanding the mapping dictionary to increase its coverage, considering all Dxx MeSH subcategories instead of being limited to the D01–D04 subset. This increase in scope resulted in improvements across all metrics, with notorious improvements being observed for F1-score (0.5417 to 0.6653) and recall (0.3819 to 0.5160), along with a slight increase in precision (0.9316 to 0.9361). The next iteration followed a similar strategy, further exploring the expansion of dictionary coverage by including mappings from the SCR file. Once again, significant improvements were observed for F1-score (0.6653 to 0.7764) and recall (0.5160 to 0.6594), while precision reached its maximum value of 0.9439.
All of the previously mentioned configurations exclusively explored external resources for the creation of the mapping dictionaries, resulting in generalizable solutions without any bias from the challenge dataset. However, despite achieving the largest mapping coverage in the dictionary from Configuration 7, it still did not manage to capture all MeSH codes present in the gold-standard annotations for the training dataset. Therefore, in the final configuration (8) we introduced prior knowledge from the training dataset into the mapping dictionary, by having the remaining unmapped entities annotated using complex mappings present in the gold-standard annotations (which were added to the mapping dictionary). To avoid information cross-talk and have a blind evaluation, during system development we only extracted complex mappings from the ‘train’ and ‘dev’ partitions of the training set and evaluated the system in the ‘test’ partition of the training set, obtaining the results presented in Table 7. As it is possible to observe, the addition of curated complex mappings from the gold-standard annotations led to a final significant performance increase, with the F1-score increasing from 0.7764 to 0.8608, recall increasing from 0.6594 to 0.7957 and precision decreasing from 0.9439 to 0.9375.
Due to its success, Configuration 8 was selected for the final entity normalization system, now adding curated concept mappings from the ‘train’, ‘dev’ and ‘test’ partitions of the training set to the mapping dictionary and evaluating the resulting system in the actual test dataset. Official challenge results for the normalization task are presented in Table 3.
Upon inspection of the rule-based system performance in the development (Table 7) and test datasets (Table 3), a clear drop in performance was noticeable when moving from development to test time, with the F1-score decreasing ∼5% points, from 0.8608 to 0.8136. Nevertheless, it is important to consider that all system performances presented in Table 7 were obtained based on the non-normalized entities provided in the gold-standard training annotations, i.e. assuming an ideal scenario where the preceding NER component would correctly detect every chemical entity in the training dataset. Since challenge submissions were based on the use of an end-to-end pipeline where errors are propagated from task to task (e.g. from entity recognition to entity normalization to chemical indexing) and there was the possibility of having new complex mappings in the test dataset that were not present in the development dataset and thus not covered in the mapping dictionary, the verified decrease in normalization performance was actually expected beforehand.
Since the ‘Annotator’ was enhanced in a post-challenge contribution, with its NER performance improving ∼3% points comparatively to our best official chemical recognition submission, and ‘Normalizer’ performance directly depends on that of the ‘Annotator’, we expected to verify performance gains in the chemical normalization task. In the left side of Table 8 we present the named entity normalization results of the rule-based approach, which was applied over the last three entity-level ensemble runs of Table 6. As suspected, by using an improved ‘Annotator’ we managed to outscore our previous best challenge results, attaining a strict F1-score of 0.8216, which demonstrates the cascading behavior of the proposed end-to-end pipeline. To evaluate the impact of the deep-learning component, we extended the previous experiment by applying the deep-learning component over the rule-based approach in order to map the remaining unmapped entities. The obtained results are presented on the right side of Table 8 and demonstrate that using the deep-learning component leads to slight yet consistent improvements over previous results, raising our overall best result to a strict F1-score of 0.8275. Another interesting result from inspecting the precision and recall metrics is that the deep-learning component increases recall at the expense of precision, which is the expected behavior since it utilizes embedding representations to find possible MeSH terms. Finally, although in our proceedings paper it was not clear whether the deep-learning component was beneficial or not for entity normalization, newly performed experiments showed that this component did indeed help our overall solution.
Comparison between the named entity normalization results, evaluated on the final NLM-Chem test set, with rule-based methods and rule-based plus the deep-learning component. Normalization was performed using each of the three ensemble models, fine-tuned in the entire NLM-Chem training set, presented in Table 6. The highest F1-score results are highlighted in bold.
| . | Rule-based . | . | Rule-based plus DL component . | ||||
|---|---|---|---|---|---|---|---|
| Rowa . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| 1 | 0.8745 | 0.7748 | 0.8216 | 0.8428 | 0.8082 | 0.8251 | |
| 2 | 0.8828 | 0.7754 | 0.8256 | 0.8494 | 0.8067 | 0.8275 | |
| 3 | 0.8801 | 0.7739 | 0.8236 | 0.8446 | 0.8045 | 0.8241 | |
| . | Rule-based . | . | Rule-based plus DL component . | ||||
|---|---|---|---|---|---|---|---|
| Rowa . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| 1 | 0.8745 | 0.7748 | 0.8216 | 0.8428 | 0.8082 | 0.8251 | |
| 2 | 0.8828 | 0.7754 | 0.8256 | 0.8494 | 0.8067 | 0.8275 | |
| 3 | 0.8801 | 0.7739 | 0.8236 | 0.8446 | 0.8045 | 0.8241 | |
a Row 1 corresponds to the entity-level ensemble model with no pre-training. Row 2 corresponds to the entity-level ensemble model pre-trained with the CDR, CHEMDNER and DrugProt datasets. Row 3 corresponds to the entity-level ensemble model that also uses the NLM-Chem-Syn data in pre-training.
Comparison between the named entity normalization results, evaluated on the final NLM-Chem test set, with rule-based methods and rule-based plus the deep-learning component. Normalization was performed using each of the three ensemble models, fine-tuned in the entire NLM-Chem training set, presented in Table 6. The highest F1-score results are highlighted in bold.
| . | Rule-based . | . | Rule-based plus DL component . | ||||
|---|---|---|---|---|---|---|---|
| Rowa . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| 1 | 0.8745 | 0.7748 | 0.8216 | 0.8428 | 0.8082 | 0.8251 | |
| 2 | 0.8828 | 0.7754 | 0.8256 | 0.8494 | 0.8067 | 0.8275 | |
| 3 | 0.8801 | 0.7739 | 0.8236 | 0.8446 | 0.8045 | 0.8241 | |
| . | Rule-based . | . | Rule-based plus DL component . | ||||
|---|---|---|---|---|---|---|---|
| Rowa . | Precision . | Recall . | F1-score . | . | Precision . | Recall . | F1-score . |
| 1 | 0.8745 | 0.7748 | 0.8216 | 0.8428 | 0.8082 | 0.8251 | |
| 2 | 0.8828 | 0.7754 | 0.8256 | 0.8494 | 0.8067 | 0.8275 | |
| 3 | 0.8801 | 0.7739 | 0.8236 | 0.8446 | 0.8045 | 0.8241 | |
a Row 1 corresponds to the entity-level ensemble model with no pre-training. Row 2 corresponds to the entity-level ensemble model pre-trained with the CDR, CHEMDNER and DrugProt datasets. Row 3 corresponds to the entity-level ensemble model that also uses the NLM-Chem-Syn data in pre-training.
Chemical indexing
The results of the chemical indexing task are directly influenced by the success of the previous tasks. Differently from the previous ones, for the chemical indexing task our system had results from official submissions and unofficial submissions (post-challenge) and also from the post-challenge extension of the work. Results for the official submissions are available in the last section of Table 3, whereas results for the best unofficial submission are presented in Table 9.
Performance comparison between participating teams in the NLM-Chem challenge. The teams and results presented here were obtained from the challenge overview paper (5). NER, normalization and indexing F1-score results, using strict evaluation, were obtained on the NLM-Chem final evaluation set. Teams are sorted according to the NER score for simplicity. The highest value in each task, considering only official results, is highlighted in bold. R: row. Norm.: normalization. Index.: indexing.
| Row . | Worka . | Models or frameworks employed . | NERb . | Normalizationb . | Indexingb . |
|---|---|---|---|---|---|
| 1 | Kim et al. (80) | Bio-LM-Large (94), BioSyn (95), SapBERT (90) | 0.8672 | 0.7831 | – |
| 2 | Erdengasileng et al. (86) | PubMedBERT (83) | 0.8600 | 0.8101 | – |
| Unofficialc | – | – | 0.4825 | ||
| 3 | Adams et al. (96) | BioMegatron (97), BioBERT (54) | 0.8571 | 0.5208 | – |
| 4 | Chiu et al. (98) | BioM-Transformers (99), PubMedBERT (83) | 0.8521 | 0.8072 | – |
| 5 | Bevan and Hodgskiss (100) | PubMedBERT (83) | 0.8493 | 0.7870 | – |
| Unofficialc | – | – | 0.3334 | ||
| 6 | Team 104 | – | 0.8463 | 0.7078 | – |
| 7 | Team 148 | – | 0.8459 | 0.7008 | – |
| 8 | Ours (78) | PubMedBERT (83), SapBERT (90) | 0.8454 | 0.8136 | 0.4664 |
| Unofficialc | – | – | 0.4651 | ||
| Post-challenged | 0.8731 | 0.8275 | 0.4849 | ||
| 9 | Team 149 | – | 0.8416 | 0.7025 | – |
| 10 | Team 146 | – | 0.8415 | 0.6744 | – |
| 11 | Tsujimura et al. (101) | SciBERT (102) | 0.8284 | 0.7954 | – |
| Unofficialc | – | – | 0.3806 | ||
| 12 | Benchmark (5) | BioBERT (54) | 0.8149 | 0.7889 | 0.4141 |
| 13 | López-Úbeda et al. (103) | ELMo (104), BioBERT (54) | 0.8136 | 0.7763 | – |
| 14 | Mercer and Alliheedi (105) | Stanza (106, 107) | 0.6492 | 0.5965 | – |
| 15 | Mobasher et al. (108) | BioBERT (54) | 0.6049 | – | – |
| Unofficialc | – | 0.4511 | – | ||
| 16 | Team 116 | – | 0.3109 | – | – |
| Row . | Worka . | Models or frameworks employed . | NERb . | Normalizationb . | Indexingb . |
|---|---|---|---|---|---|
| 1 | Kim et al. (80) | Bio-LM-Large (94), BioSyn (95), SapBERT (90) | 0.8672 | 0.7831 | – |
| 2 | Erdengasileng et al. (86) | PubMedBERT (83) | 0.8600 | 0.8101 | – |
| Unofficialc | – | – | 0.4825 | ||
| 3 | Adams et al. (96) | BioMegatron (97), BioBERT (54) | 0.8571 | 0.5208 | – |
| 4 | Chiu et al. (98) | BioM-Transformers (99), PubMedBERT (83) | 0.8521 | 0.8072 | – |
| 5 | Bevan and Hodgskiss (100) | PubMedBERT (83) | 0.8493 | 0.7870 | – |
| Unofficialc | – | – | 0.3334 | ||
| 6 | Team 104 | – | 0.8463 | 0.7078 | – |
| 7 | Team 148 | – | 0.8459 | 0.7008 | – |
| 8 | Ours (78) | PubMedBERT (83), SapBERT (90) | 0.8454 | 0.8136 | 0.4664 |
| Unofficialc | – | – | 0.4651 | ||
| Post-challenged | 0.8731 | 0.8275 | 0.4849 | ||
| 9 | Team 149 | – | 0.8416 | 0.7025 | – |
| 10 | Team 146 | – | 0.8415 | 0.6744 | – |
| 11 | Tsujimura et al. (101) | SciBERT (102) | 0.8284 | 0.7954 | – |
| Unofficialc | – | – | 0.3806 | ||
| 12 | Benchmark (5) | BioBERT (54) | 0.8149 | 0.7889 | 0.4141 |
| 13 | López-Úbeda et al. (103) | ELMo (104), BioBERT (54) | 0.8136 | 0.7763 | – |
| 14 | Mercer and Alliheedi (105) | Stanza (106, 107) | 0.6492 | 0.5965 | – |
| 15 | Mobasher et al. (108) | BioBERT (54) | 0.6049 | – | – |
| Unofficialc | – | 0.4511 | – | ||
| 16 | Team 116 | – | 0.3109 | – | – |
a Teams that did not submit a description paper for the BioCreative VII workshop are denoted with their team numbers according to the challenge overview paper (5). The benchmark shows the performance of a baseline model shared by the challenge organizers.
b Note that the NER, normalization and indexing results may not correspond to the same model run. We only present the highest obtained results of each team, but teams were allowed to submit several runs.
c Additional submissions were allowed posterior to the challenge, although not being considered in the official challenge results.
d Our improved results due to post-challenge enhancements reported in this manuscript.
Performance comparison between participating teams in the NLM-Chem challenge. The teams and results presented here were obtained from the challenge overview paper (5). NER, normalization and indexing F1-score results, using strict evaluation, were obtained on the NLM-Chem final evaluation set. Teams are sorted according to the NER score for simplicity. The highest value in each task, considering only official results, is highlighted in bold. R: row. Norm.: normalization. Index.: indexing.
| Row . | Worka . | Models or frameworks employed . | NERb . | Normalizationb . | Indexingb . |
|---|---|---|---|---|---|
| 1 | Kim et al. (80) | Bio-LM-Large (94), BioSyn (95), SapBERT (90) | 0.8672 | 0.7831 | – |
| 2 | Erdengasileng et al. (86) | PubMedBERT (83) | 0.8600 | 0.8101 | – |
| Unofficialc | – | – | 0.4825 | ||
| 3 | Adams et al. (96) | BioMegatron (97), BioBERT (54) | 0.8571 | 0.5208 | – |
| 4 | Chiu et al. (98) | BioM-Transformers (99), PubMedBERT (83) | 0.8521 | 0.8072 | – |
| 5 | Bevan and Hodgskiss (100) | PubMedBERT (83) | 0.8493 | 0.7870 | – |
| Unofficialc | – | – | 0.3334 | ||
| 6 | Team 104 | – | 0.8463 | 0.7078 | – |
| 7 | Team 148 | – | 0.8459 | 0.7008 | – |
| 8 | Ours (78) | PubMedBERT (83), SapBERT (90) | 0.8454 | 0.8136 | 0.4664 |
| Unofficialc | – | – | 0.4651 | ||
| Post-challenged | 0.8731 | 0.8275 | 0.4849 | ||
| 9 | Team 149 | – | 0.8416 | 0.7025 | – |
| 10 | Team 146 | – | 0.8415 | 0.6744 | – |
| 11 | Tsujimura et al. (101) | SciBERT (102) | 0.8284 | 0.7954 | – |
| Unofficialc | – | – | 0.3806 | ||
| 12 | Benchmark (5) | BioBERT (54) | 0.8149 | 0.7889 | 0.4141 |
| 13 | López-Úbeda et al. (103) | ELMo (104), BioBERT (54) | 0.8136 | 0.7763 | – |
| 14 | Mercer and Alliheedi (105) | Stanza (106, 107) | 0.6492 | 0.5965 | – |
| 15 | Mobasher et al. (108) | BioBERT (54) | 0.6049 | – | – |
| Unofficialc | – | 0.4511 | – | ||
| 16 | Team 116 | – | 0.3109 | – | – |
| Row . | Worka . | Models or frameworks employed . | NERb . | Normalizationb . | Indexingb . |
|---|---|---|---|---|---|
| 1 | Kim et al. (80) | Bio-LM-Large (94), BioSyn (95), SapBERT (90) | 0.8672 | 0.7831 | – |
| 2 | Erdengasileng et al. (86) | PubMedBERT (83) | 0.8600 | 0.8101 | – |
| Unofficialc | – | – | 0.4825 | ||
| 3 | Adams et al. (96) | BioMegatron (97), BioBERT (54) | 0.8571 | 0.5208 | – |
| 4 | Chiu et al. (98) | BioM-Transformers (99), PubMedBERT (83) | 0.8521 | 0.8072 | – |
| 5 | Bevan and Hodgskiss (100) | PubMedBERT (83) | 0.8493 | 0.7870 | – |
| Unofficialc | – | – | 0.3334 | ||
| 6 | Team 104 | – | 0.8463 | 0.7078 | – |
| 7 | Team 148 | – | 0.8459 | 0.7008 | – |
| 8 | Ours (78) | PubMedBERT (83), SapBERT (90) | 0.8454 | 0.8136 | 0.4664 |
| Unofficialc | – | – | 0.4651 | ||
| Post-challenged | 0.8731 | 0.8275 | 0.4849 | ||
| 9 | Team 149 | – | 0.8416 | 0.7025 | – |
| 10 | Team 146 | – | 0.8415 | 0.6744 | – |
| 11 | Tsujimura et al. (101) | SciBERT (102) | 0.8284 | 0.7954 | – |
| Unofficialc | – | – | 0.3806 | ||
| 12 | Benchmark (5) | BioBERT (54) | 0.8149 | 0.7889 | 0.4141 |
| 13 | López-Úbeda et al. (103) | ELMo (104), BioBERT (54) | 0.8136 | 0.7763 | – |
| 14 | Mercer and Alliheedi (105) | Stanza (106, 107) | 0.6492 | 0.5965 | – |
| 15 | Mobasher et al. (108) | BioBERT (54) | 0.6049 | – | – |
| Unofficialc | – | 0.4511 | – | ||
| 16 | Team 116 | – | 0.3109 | – | – |
a Teams that did not submit a description paper for the BioCreative VII workshop are denoted with their team numbers according to the challenge overview paper (5). The benchmark shows the performance of a baseline model shared by the challenge organizers.
b Note that the NER, normalization and indexing results may not correspond to the same model run. We only present the highest obtained results of each team, but teams were allowed to submit several runs.
c Additional submissions were allowed posterior to the challenge, although not being considered in the official challenge results.
d Our improved results due to post-challenge enhancements reported in this manuscript.
The original rule-based approach obtained strict F1-scores of 0.4664 and 0.4437, outperforming the benchmark solution and attaining the top performance in official challenge submissions, whereas the TF-IDF solution achieved strict F1-scores below 0.4, failing to perform even on par with the benchmark. Due to the overall poor performance of the latter approach, TF-IDF was sidelined and improvement efforts were focused on the Indexer’s rule-based system, where a new rule was introduced for evaluating MeSH codes present in the conclusion section. Previously existing rules were slightly adjusted, and all occurrence thresholds were modified to reflect the integration of a novel rule in the system. Despite presenting significant and consistent improvements of ∼5% points across all three training dataset partitions (‘train’, ‘dev’ and ‘test’), the enhanced rule-based system showed a decrease in strict F1-score in unofficial submissions (0.4664 to 0.4651).
Posterior to the challenge and due to the contradictory and unexpected behavior of the enhanced rule-based system in unofficial submissions, a deeper analysis was performed on this system to evaluate the impact of each rule as well as the impact of varying occurrence thresholds. Here, the ‘train’ and ‘dev’ partitions of the training dataset were merged and used as a development dataset, while the ‘test’ partition was used for evaluation.
The selected rules were not defined using any methodical approach due to the sheer number of existing structural elements within each scientific manuscript. For instance, a simple element such as a caption could be referenced using different tags depending on its origin (e.g. table, figure and supplementary material, among others), which increased the number of options to be considered. Therefore, in an initial stage, we analyzed all these elements and experimented using all of them in the searching range. However, the initial results were poor and led us to reduce the type of elements to consider in the indexing mechanism. Although sections such as methods, results and discussion could have the actual MeSH codes of interest, we discovered that these sections led to the detection of numerous noisy annotations, which negatively influenced the indexing performance of the system. Therefore, we abandoned these and focused only on the elements of the manuscripts that have more emphasis, leading to the selection of the title, abstract, conclusions and all captions from tables and figures. By establishing these sections as the searching scope for MeSH codes, we started to identify weights for each section to define whether the annotated MeSH should be indexed or not.
In this analysis we focused on assessing the indexing potential of using each rule independently. For that, rules for the caption, abstract, title and conclusion sections were used separately on the development dataset, varying their corresponding occurrence thresholds in a range between 1% and 30%, in increments of 1%. The obtained results demonstrated two main trends: firstly, every rule had better performances when using smaller thresholds, with thresholds >10% leading to a rapid decay in indexing performance; and secondly, the rule for the conclusion section obtained significantly worse results than the remaining ones, showing that this section of full-text papers contains a smaller amount of relevant MeSH codes. For the sake of simplicity and owing to the first identified trend, Table 10 reports the obtained results for a threshold range between 1% and 10%.
Results obtained on an NLM-Chem development dataset (‘train’+‘dev’ subsets) by indexing the documents using each rule independently. For simplicity purposes, we only report results for the threshold variation interval of 1–10% as this yielded the best performances. The highest F1-score results are highlighted in bold.
| Threshold (%) . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Rule for MeSH codes in captions | |||
| 1 | 0.2369 | 0.6196 | 0.3427 |
| 2 | 0.2844 | 0.6000 | 0.3859 |
| 3 | 0.3274 | 0.5804 | 0.4187 |
| 4 | 0.3684 | 0.5490 | 0.4409 |
| 5 | 0.3900 | 0.5216 | 0.4463 |
| 6 | 0.4262 | 0.4980 | 0.4593 |
| 7 | 0.4457 | 0.4667 | 0.4559 |
| 8 | 0.4615 | 0.4471 | 0.4542 |
| 9 | 0.4758 | 0.4235 | 0.4481 |
| 10 | 0.5025 | 0.3961 | 0.4430 |
| Rule for MeSH codes in the abstract | |||
| 1 | 0.3264 | 0.6784 | 0.4408 |
| 2 | 0.3573 | 0.6431 | 0.4594 |
| 3 | 0.3873 | 0.6196 | 0.4766 |
| 4 | 0.4048 | 0.5922 | 0.4809 |
| 5 | 0.4128 | 0.5569 | 0.4741 |
| 6 | 0.4363 | 0.5373 | 0.4815 |
| 7 | 0.4429 | 0.5020 | 0.4706 |
| 8 | 0.4636 | 0.4745 | 0.4690 |
| 9 | 0.4793 | 0.4549 | 0.4668 |
| 10 | 0.5023 | 0.4196 | 0.4573 |
| Rule for MeSH codes in the title | |||
| 1 | 0.5066 | 0.4549 | 0.4793 |
| 2 | 0.5068 | 0.4392 | 0.4706 |
| 3 | 0.5140 | 0.4314 | 0.4691 |
| 4 | 0.5222 | 0.4157 | 0.4629 |
| 5 | 0.5282 | 0.4039 | 0.4578 |
| 6 | 0.5538 | 0.4039 | 0.4671 |
| 7 | 0.5568 | 0.3843 | 0.4548 |
| 8 | 0.5509 | 0.3608 | 0.4360 |
| 9 | 0.5605 | 0.3451 | 0.4272 |
| 10 | 0.5664 | 0.3176 | 0.4070 |
| Rule for MeSH codes in the conclusions | |||
| 1 | 0.5000 | 0.0745 | 0.1297 |
| 2 | 0.5152 | 0.0667 | 0.1181 |
| 3 | 0.6296 | 0.0667 | 0.1206 |
| 4 | 0.6538 | 0.0667 | 0.1210 |
| 5 | 0.6400 | 0.0627 | 0.1143 |
| 6 | 0.6667 | 0.0627 | 0.1147 |
| 7 | 0.6364 | 0.0549 | 0.1011 |
| 8 | 0.6364 | 0.0549 | 0.1011 |
| 9 | 0.6842 | 0.0510 | 0.0949 |
| 10 | 0.8000 | 0.0471 | 0.0889 |
| Threshold (%) . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Rule for MeSH codes in captions | |||
| 1 | 0.2369 | 0.6196 | 0.3427 |
| 2 | 0.2844 | 0.6000 | 0.3859 |
| 3 | 0.3274 | 0.5804 | 0.4187 |
| 4 | 0.3684 | 0.5490 | 0.4409 |
| 5 | 0.3900 | 0.5216 | 0.4463 |
| 6 | 0.4262 | 0.4980 | 0.4593 |
| 7 | 0.4457 | 0.4667 | 0.4559 |
| 8 | 0.4615 | 0.4471 | 0.4542 |
| 9 | 0.4758 | 0.4235 | 0.4481 |
| 10 | 0.5025 | 0.3961 | 0.4430 |
| Rule for MeSH codes in the abstract | |||
| 1 | 0.3264 | 0.6784 | 0.4408 |
| 2 | 0.3573 | 0.6431 | 0.4594 |
| 3 | 0.3873 | 0.6196 | 0.4766 |
| 4 | 0.4048 | 0.5922 | 0.4809 |
| 5 | 0.4128 | 0.5569 | 0.4741 |
| 6 | 0.4363 | 0.5373 | 0.4815 |
| 7 | 0.4429 | 0.5020 | 0.4706 |
| 8 | 0.4636 | 0.4745 | 0.4690 |
| 9 | 0.4793 | 0.4549 | 0.4668 |
| 10 | 0.5023 | 0.4196 | 0.4573 |
| Rule for MeSH codes in the title | |||
| 1 | 0.5066 | 0.4549 | 0.4793 |
| 2 | 0.5068 | 0.4392 | 0.4706 |
| 3 | 0.5140 | 0.4314 | 0.4691 |
| 4 | 0.5222 | 0.4157 | 0.4629 |
| 5 | 0.5282 | 0.4039 | 0.4578 |
| 6 | 0.5538 | 0.4039 | 0.4671 |
| 7 | 0.5568 | 0.3843 | 0.4548 |
| 8 | 0.5509 | 0.3608 | 0.4360 |
| 9 | 0.5605 | 0.3451 | 0.4272 |
| 10 | 0.5664 | 0.3176 | 0.4070 |
| Rule for MeSH codes in the conclusions | |||
| 1 | 0.5000 | 0.0745 | 0.1297 |
| 2 | 0.5152 | 0.0667 | 0.1181 |
| 3 | 0.6296 | 0.0667 | 0.1206 |
| 4 | 0.6538 | 0.0667 | 0.1210 |
| 5 | 0.6400 | 0.0627 | 0.1143 |
| 6 | 0.6667 | 0.0627 | 0.1147 |
| 7 | 0.6364 | 0.0549 | 0.1011 |
| 8 | 0.6364 | 0.0549 | 0.1011 |
| 9 | 0.6842 | 0.0510 | 0.0949 |
| 10 | 0.8000 | 0.0471 | 0.0889 |
Results obtained on an NLM-Chem development dataset (‘train’+‘dev’ subsets) by indexing the documents using each rule independently. For simplicity purposes, we only report results for the threshold variation interval of 1–10% as this yielded the best performances. The highest F1-score results are highlighted in bold.
| Threshold (%) . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Rule for MeSH codes in captions | |||
| 1 | 0.2369 | 0.6196 | 0.3427 |
| 2 | 0.2844 | 0.6000 | 0.3859 |
| 3 | 0.3274 | 0.5804 | 0.4187 |
| 4 | 0.3684 | 0.5490 | 0.4409 |
| 5 | 0.3900 | 0.5216 | 0.4463 |
| 6 | 0.4262 | 0.4980 | 0.4593 |
| 7 | 0.4457 | 0.4667 | 0.4559 |
| 8 | 0.4615 | 0.4471 | 0.4542 |
| 9 | 0.4758 | 0.4235 | 0.4481 |
| 10 | 0.5025 | 0.3961 | 0.4430 |
| Rule for MeSH codes in the abstract | |||
| 1 | 0.3264 | 0.6784 | 0.4408 |
| 2 | 0.3573 | 0.6431 | 0.4594 |
| 3 | 0.3873 | 0.6196 | 0.4766 |
| 4 | 0.4048 | 0.5922 | 0.4809 |
| 5 | 0.4128 | 0.5569 | 0.4741 |
| 6 | 0.4363 | 0.5373 | 0.4815 |
| 7 | 0.4429 | 0.5020 | 0.4706 |
| 8 | 0.4636 | 0.4745 | 0.4690 |
| 9 | 0.4793 | 0.4549 | 0.4668 |
| 10 | 0.5023 | 0.4196 | 0.4573 |
| Rule for MeSH codes in the title | |||
| 1 | 0.5066 | 0.4549 | 0.4793 |
| 2 | 0.5068 | 0.4392 | 0.4706 |
| 3 | 0.5140 | 0.4314 | 0.4691 |
| 4 | 0.5222 | 0.4157 | 0.4629 |
| 5 | 0.5282 | 0.4039 | 0.4578 |
| 6 | 0.5538 | 0.4039 | 0.4671 |
| 7 | 0.5568 | 0.3843 | 0.4548 |
| 8 | 0.5509 | 0.3608 | 0.4360 |
| 9 | 0.5605 | 0.3451 | 0.4272 |
| 10 | 0.5664 | 0.3176 | 0.4070 |
| Rule for MeSH codes in the conclusions | |||
| 1 | 0.5000 | 0.0745 | 0.1297 |
| 2 | 0.5152 | 0.0667 | 0.1181 |
| 3 | 0.6296 | 0.0667 | 0.1206 |
| 4 | 0.6538 | 0.0667 | 0.1210 |
| 5 | 0.6400 | 0.0627 | 0.1143 |
| 6 | 0.6667 | 0.0627 | 0.1147 |
| 7 | 0.6364 | 0.0549 | 0.1011 |
| 8 | 0.6364 | 0.0549 | 0.1011 |
| 9 | 0.6842 | 0.0510 | 0.0949 |
| 10 | 0.8000 | 0.0471 | 0.0889 |
| Threshold (%) . | Precision . | Recall . | F1-Score . |
|---|---|---|---|
| Rule for MeSH codes in captions | |||
| 1 | 0.2369 | 0.6196 | 0.3427 |
| 2 | 0.2844 | 0.6000 | 0.3859 |
| 3 | 0.3274 | 0.5804 | 0.4187 |
| 4 | 0.3684 | 0.5490 | 0.4409 |
| 5 | 0.3900 | 0.5216 | 0.4463 |
| 6 | 0.4262 | 0.4980 | 0.4593 |
| 7 | 0.4457 | 0.4667 | 0.4559 |
| 8 | 0.4615 | 0.4471 | 0.4542 |
| 9 | 0.4758 | 0.4235 | 0.4481 |
| 10 | 0.5025 | 0.3961 | 0.4430 |
| Rule for MeSH codes in the abstract | |||
| 1 | 0.3264 | 0.6784 | 0.4408 |
| 2 | 0.3573 | 0.6431 | 0.4594 |
| 3 | 0.3873 | 0.6196 | 0.4766 |
| 4 | 0.4048 | 0.5922 | 0.4809 |
| 5 | 0.4128 | 0.5569 | 0.4741 |
| 6 | 0.4363 | 0.5373 | 0.4815 |
| 7 | 0.4429 | 0.5020 | 0.4706 |
| 8 | 0.4636 | 0.4745 | 0.4690 |
| 9 | 0.4793 | 0.4549 | 0.4668 |
| 10 | 0.5023 | 0.4196 | 0.4573 |
| Rule for MeSH codes in the title | |||
| 1 | 0.5066 | 0.4549 | 0.4793 |
| 2 | 0.5068 | 0.4392 | 0.4706 |
| 3 | 0.5140 | 0.4314 | 0.4691 |
| 4 | 0.5222 | 0.4157 | 0.4629 |
| 5 | 0.5282 | 0.4039 | 0.4578 |
| 6 | 0.5538 | 0.4039 | 0.4671 |
| 7 | 0.5568 | 0.3843 | 0.4548 |
| 8 | 0.5509 | 0.3608 | 0.4360 |
| 9 | 0.5605 | 0.3451 | 0.4272 |
| 10 | 0.5664 | 0.3176 | 0.4070 |
| Rule for MeSH codes in the conclusions | |||
| 1 | 0.5000 | 0.0745 | 0.1297 |
| 2 | 0.5152 | 0.0667 | 0.1181 |
| 3 | 0.6296 | 0.0667 | 0.1206 |
| 4 | 0.6538 | 0.0667 | 0.1210 |
| 5 | 0.6400 | 0.0627 | 0.1143 |
| 6 | 0.6667 | 0.0627 | 0.1147 |
| 7 | 0.6364 | 0.0549 | 0.1011 |
| 8 | 0.6364 | 0.0549 | 0.1011 |
| 9 | 0.6842 | 0.0510 | 0.0949 |
| 10 | 0.8000 | 0.0471 | 0.0889 |
Since using each rule in an individual manner resulted in an indexing system that could not index at least one MeSH code in every document, we performed another experiment to assess the maximum number of documents where each rule could index at least one MeSH code. For that, each rule was used in the development dataset with a lenient occurrence threshold of 1%. The obtained results showed that the rule for captions was able to index codes in 93% of the documents, the rule for abstracts managed to index codes in 98% of the documents, the rule for titles managed 87% and finally the rule for the conclusion section only achieved a reduced portion of 10%.
For the final experiment, we assessed the influence of using different rule combinations in indexing performance while seeking for an optimal rule set and its corresponding occurrence thresholds. Here, the rule-based system was evaluated using every possible combination of two and three rules and finally using all four rules. In every test the occurrence threshold for each rule was varied between 1% and 30%, resulting in an exponentially sized search space. The best indexing performance achieved on the development dataset was obtained using all four rules with occurrence thresholds of 2%, 22%, 10% and 10% for the title, captions, abstract and conclusion rules, respectively. The resulting configuration was evaluated on the ‘test’ split of the training dataset, obtaining a precision of 0.4328, recall of 0.5321 and F1-score of 0.4774. Finally, this chemical indexing system was evaluated on the best NER + Normalization run on the test dataset, managing to index 95.5% of the documents and obtaining a precision of 0.5017, recall of 0.4691 and strict F1-score of 0.4849, surpassing the best indexing performance among all submissions (both official and unofficial) in this task of the NLM-Chem BioCreative VII challenge track.
Finally, for contextualization purposes Table 9 shows a performance comparison between all participating teams in the NLM-Chem challenge, including our original submissions and our best post-challenge runs.
Error analysis
An error analysis was conducted focusing on the first module of the proposed pipeline, during which it was possible to identify situations where the NER model wrongfully predicted false-positive chemical mentions and cases where it failed to identify the correct annotations. In this section, we not only enumerate and discuss some possible causes for verified entity recognition errors, but also evaluate the impact of error propagation in the performance of the cascaded pipeline.
Beginning with tokenization, this process hinders NER for entities that do not respect tokenization boundaries. This error occurred in 156 entities from the 38 339 entities present in the challenge corpora, with two examples being presented in Figure 5. Overall, this may be alleviated by tokenization-free models or using string-pattern match methods in a post-processing phase.

Two examples of entity recognition errors due to tokenization boundaries. Blue boxes represent gold standard entity annotations, and the yellow boxes highlight characters that cannot be disentangled from the true annotations due to tokenization. The first error (a) occurs in the document PMC 3 661 362, where Na+ and K+ are gold-standard entity mentions. However, the PubMedBERT tokenizer produces the tokens INa and IK, which makes it impossible to correctly predict Na+ and K+ using a token-level tagging schema. The ‘closest’ predictions would be INa+ and IK+, which would be incorrect according to strict evaluation. The second example (b) was extracted from PMC 2 952 795 containing a similar issue with the term Ocarboxylate, where Oc forms a single token.
Furthermore, in the manual process of annotating corpora it is acceptable that even expert curators may make some errors unintentionally or that they disagree in some specific annotations, as shown in Figure 6. Even with specific guidelines, experts can have different interpretations, which is the reason we hypothesize that gold-standard annotations may contain a small number of partially incorrect annotations (for example, an incorrect span), as presented in Figure 7.

An example of a potential inconsistent annotation found in the document PMC 2 254 971. In the first sentence (a), the curators considered propidium iodide as a chemical as well as its short form PI. However, as shown with the yellow box, the term PI in the second sentence (b) was not annotated, which made us suspect if it was a missed annotation.

Example of an annotation with incorrect span from document PMC 5 096 026. The term polypyridy was mistakenly annotated instead of polypyridyl, since the last character was not included in the entity mention span.
Another problem that influences entity detection is related with limited instance representation. In some cases, the 512 tokens from the BERT model are not enough, and more context (in limit, the whole document) is required to correctly perceive the surrounding context of chemicals. This limitation aggravates the tagging inconsistency problem (92), which means having annotations that are not consistent along the document, and is the source of some errors as also reported by Kim et al. (80). For example, the chemical entity ‘ST-4’ present in the PMC 555 756 article was only detected twice by our model, despite appearing five times in the gold-standard annotations of the article (Figure 8).
Example of the tagging inconsistency problem from the document PMC 555 756. Our model correctly predicted the entity ST-4 for Sentences (b) and (d), but failed to predict the remainder ST-4 mentions in Sentences (a), (c) and (e), which renders a final document annotation that appears to be contradictory.
Another specific error that our model is likely to produce is that chemical mentions may be annotated even if they are within protein mentions, which according to the annotation guidelines is undesired. For instance, in Figure 9, ‘cholesterol’ was correctly recognized by the model in one of the cases, but it is incorrectly identified in the other case since the same term belongs to a protein mention. In the future, to take into account these cases we would suggest to include an additional step for identifying protein entities.

Example of a chemical mention embedded in a protein mention from document PMC 2 096 715. Our model predicted cholesterol on both sentences as an chemical entity. However, in Sentence (a) the word cholesterol should not be identified because, according to the annotation guidelines, the chemical term appears within a protein mention.
Complex terms require expert interpretation, which for deep-learning models may not be available without extensive external knowledge. In a similar manner, new terms introduced in literature may not be easily identified by such models without having some kind of external knowledge. Such behavior holds especially true in a field where new chemicals are often being discovered and specific names are created for these new entities.
Finally, we evaluated the impact of error propagation in the complete system, which is inherent to the system due to its conception as a cascaded pipeline. Having a top-performing NER module is key to the success of the pipeline since it defines which textual entities should be forwarded to the normalization and indexing modules; thus, a great effort was placed into the development and analysis of the NER module. However, due to the existence of gold-standard annotations for the normalization and indexing subtasks, it was possible to develop and evaluate both modules based on an ‘optimal’ scenario where the NER module correctly identifies every textual entity. This assumption naturally biases system performance, boosting normalization and indexing performances during system development and leading to a more significant drop in performance during test time.
To capture the impact of error propagation as expected from a real scenario, we ran the full pipeline on the test split of the training dataset, with the obtained results being presented in Table 11. For comparison purposes, we provide system performances using gold-standard annotations versus system annotations for the normalization and indexing modules, along with the relative change in performance resultant from using system outputs instead of gold-standard annotations. Upon inspection of obtained results it is possible to observe two major trends. Firstly, there clearly exists error propagation through the pipeline, as evidenced by the performance loss in F1-score of nearly 6% and 3% in normalization and indexing, respectively. Secondly, system performances obtained when using generated annotations (Table 11) in the test split of the training dataset are close to the results reported for the full test dataset (Table 9), which demonstrates the robustness of the herein proposed system.
Performance comparison for the normalization and indexing subtasks in the ‘test’ subset from the NLM-Chem training dataset when using gold-standard annotation files versus system annotations from the previous module in the pipeline. Performance for the NER module in the same dataset is also provided to demonstrate the performance drop, resulting from loss propagation through the cascaded pipeline. The reported delta values correspond to the relative change in performance when using gold standard annotations versus system annotations.
| . | Precision . | . | Recall . | . | F1-Score . | . |
|---|---|---|---|---|---|---|
| Chemical mention recognition | ||||||
| – | 0.8685 | 0.8906 | 0.8794 | |||
| Chemical normalization to MeSH IDs | ||||||
| Gold-standard annotations | 0.9090 | 0.8366 | 0.8713 | |||
| System annotations (NER) | 0.8393 | Δ –7.67% | 0.7999 | Δ –4.39% | 0.8191 | Δ –5.99% |
| Chemical indexing | ||||||
| Gold-standard annotations | 0.4286 | 0.5229 | 0.4711 | |||
| System annotations (Normalization) | 0.4252 | Δ –0.79% | 0.4954 | Δ –5.26% | 0.4576 | Δ –2.87% |
| . | Precision . | . | Recall . | . | F1-Score . | . |
|---|---|---|---|---|---|---|
| Chemical mention recognition | ||||||
| – | 0.8685 | 0.8906 | 0.8794 | |||
| Chemical normalization to MeSH IDs | ||||||
| Gold-standard annotations | 0.9090 | 0.8366 | 0.8713 | |||
| System annotations (NER) | 0.8393 | Δ –7.67% | 0.7999 | Δ –4.39% | 0.8191 | Δ –5.99% |
| Chemical indexing | ||||||
| Gold-standard annotations | 0.4286 | 0.5229 | 0.4711 | |||
| System annotations (Normalization) | 0.4252 | Δ –0.79% | 0.4954 | Δ –5.26% | 0.4576 | Δ –2.87% |
Performance comparison for the normalization and indexing subtasks in the ‘test’ subset from the NLM-Chem training dataset when using gold-standard annotation files versus system annotations from the previous module in the pipeline. Performance for the NER module in the same dataset is also provided to demonstrate the performance drop, resulting from loss propagation through the cascaded pipeline. The reported delta values correspond to the relative change in performance when using gold standard annotations versus system annotations.
| . | Precision . | . | Recall . | . | F1-Score . | . |
|---|---|---|---|---|---|---|
| Chemical mention recognition | ||||||
| – | 0.8685 | 0.8906 | 0.8794 | |||
| Chemical normalization to MeSH IDs | ||||||
| Gold-standard annotations | 0.9090 | 0.8366 | 0.8713 | |||
| System annotations (NER) | 0.8393 | Δ –7.67% | 0.7999 | Δ –4.39% | 0.8191 | Δ –5.99% |
| Chemical indexing | ||||||
| Gold-standard annotations | 0.4286 | 0.5229 | 0.4711 | |||
| System annotations (Normalization) | 0.4252 | Δ –0.79% | 0.4954 | Δ –5.26% | 0.4576 | Δ –2.87% |
| . | Precision . | . | Recall . | . | F1-Score . | . |
|---|---|---|---|---|---|---|
| Chemical mention recognition | ||||||
| – | 0.8685 | 0.8906 | 0.8794 | |||
| Chemical normalization to MeSH IDs | ||||||
| Gold-standard annotations | 0.9090 | 0.8366 | 0.8713 | |||
| System annotations (NER) | 0.8393 | Δ –7.67% | 0.7999 | Δ –4.39% | 0.8191 | Δ –5.99% |
| Chemical indexing | ||||||
| Gold-standard annotations | 0.4286 | 0.5229 | 0.4711 | |||
| System annotations (Normalization) | 0.4252 | Δ –0.79% | 0.4954 | Δ –5.26% | 0.4576 | Δ –2.87% |
Conclusions and future work
This paper describes deep-learning and rule-based strategies for chemical entity recognition, normalization and indexing following our participation in the NLM-Chem track of the BioCreative VII challenge (Track 2). We presented in detail the post-challenge experiments we conducted and how these improved our final system performances. More precisely, we improved our NER F1-score from 0.8454 to 0.8731; NEN F1-score from 0.8136 to 0.8275 and indexing F1-score from 0.4664 to 0.4849.
For future work, we aim to tackle the tagging inconsistency problem improving the already-started document-level agreement mechanism. Additionally, to deal with the tokenization boundary problems, we believe that using tokenization-free models, such as ByT5 (93), may mitigate this problem. Regarding the normalization, we also view the added value in expanding the deep-learning component by training a dense retrieval based on the SapBERT embeddings (90) for the task of entity linking. Finally, for the indexing we intend to explore the MeSH tree structure, which may help to identify parent MeSH codes that are meaningful for indexing, but did not appear in the document.
Acknowledgement
We thank the organizers of the BioCreative VII NLM-Chem track, the authors of PubMedBERT for making their model publicly available and the reviewers for the valuable insights. We also thank Rui Lebre for helping us with the online storage for our pre-trained models and supplementary data and Hélder Brandão Antunes for improving the design of some images in the paper.
Funding
Portuguese national funds through the Foundation for Science and Technology (FCT) (2020.05784BD to T.A., SFRH/BD/137000/2018 to R.A., PD/BD/142878/2018 to J.F.S., SFRH/BD/147837/2019 to J.R.A.); national funds through the FCT (UIDB/00127/2020).



