-
PDF
- Split View
-
Views
-
Cite
Cite
Sheng-Jie Lin, Wen-Chao Yeh, Yu-Wen Chiu, Yung-Chun Chang, Min-Huei Hsu, Yi-Shin Chen, Wen-Lian Hsu, A BERT-based ensemble learning approach for the BioCreative VII challenges: full-text chemical identification and multi-label classification in PubMed articles, Database, Volume 2022, 2022, baac056, https://doi.org/10.1093/database/baac056
Close - Share Icon Share
Abstract
In this research, we explored various state-of-the-art biomedical-specific pre-trained Bidirectional Encoder Representations from Transformers (BERT) models for the National Library of Medicine - Chemistry (NLM CHEM) and LitCovid tracks in the BioCreative VII Challenge, and propose a BERT-based ensemble learning approach to integrate the advantages of various models to improve the system’s performance. The experimental results of the NLM-CHEM track demonstrate that our method can achieve remarkable performance, with F1-scores of 85% and 91.8% in strict and approximate evaluations, respectively. Moreover, the proposed Medical Subject Headings identifier (MeSH ID) normalization algorithm is effective in entity normalization, which achieved a F1-score of about 80% in both strict and approximate evaluations. For the LitCovid track, the proposed method is also effective in detecting topics in the Coronavirus disease 2019 (COVID-19) literature, which outperformed the compared methods and achieve state-of-the-art performance in the LitCovid corpus.
Database URL: https://www.ncbi.nlm.nih.gov/research/coronavirus/.
Introduction
Artificial intelligence (AI), one of the fastest-growing technologies in research, has garnered substantial investment in recent years. According to the ‘Artificial Intelligence Index Report 2021’ (1), medical fields have received more than USD 13.8 billion in private AI investment, which is 4.5 times higher than in 2019. In particular, COVID-19 has had an impact on AI development, such as the adoption of machine learning techniques to accelerate COVID-related drug discovery. Furthermore, a vast amount of medical textual data exists in the public domain, such as on social media, online forums or in published articles, and this data includes patients’ clinical notes and biological publications (2). These text-based data are growing rapidly and can offer valuable insights with the help of text mining (3). However, most text data exist as low-quality and unstructured data. For this reason, Natural Language Processing (NLP) is seen as a bridge between human language and computers, enabling machines to understand, process and analyze human language (4). NLP’s significance as a tool aiding comprehension of human-generated data is a logical consequence of the context-dependency of data. Data becomes more meaningful when its context is better understood, which makes text analysis and mining easier (5). Therefore, NLP methods aid in the examination of a large amount of unstructured and low-quality text and the discovery of relevant insights (6), and NLP methods are frequently utilized for this purpose.
Taking the example of biomedical research, the number of publications in electronic format that can be accessed online is growing rapidly as a result of the swift advancement of technology. For instance, PubMed contains more than 33 million articles and is growing by more than 1000 articles per day (7). With such rapid explosion of new information, it is impossible for readers to keep up-to-date with all the relevant research. As a result, automatic knowledge mining and distillation techniques of the biomedical literature have become more prevalent. Biomedical literature mining (BLM), which uses NLP and/or text mining techniques, has gained prominence consequentially. In view of the importance of BLM and the lack of common standards or shared evaluation criteria to enable comparison among the different approaches, the BioCreative (http://www.biocreative.org/) (Critical Assessment of Information Extraction in Biology) organization was established. This organization hosts an annual Challenge to evaluate text mining and information extraction systems that are applied to biological and biochemical domains. The Challenge and the accompanying BioCreative Workshops promote interactions between the text mining and biomedical communities and facilitate the development of new applications as well as improvements to existing text mining systems to satisfy key research needs. The results have been novel applications that can assist in the knowledge discovery process.
To efficiently extract knowledge from biomedical literature, we can perform two fundamental tasks: (i) biomedical named entity recognition (BioNER) and normalization and (ii) biomedical literature classification. These tasks are explained in detail as follows. In this paper, we present a Bidirectional Encoder Representations from Transformers (BERT)-based ensemble learning approach for Track 2 and Track 5 in the BioCreative VII Challenge. For Track 2 (NLM-CHEM track: Full-text Chemical Identification and Indexing in PubMed articles), we integrate different BERT models through ensemble learning for recognizing chemical entities. We also tackle the entity linking problem in chemical normalization using a dynamic programming algorithm. Our framework was shown to surpass benchmarks as well as the median of the compared methods. Furthermore, for Track 5 (LitCovid track: multi-label topic classification for COVID-19 literature annotation), our BERT-based ensemble learning method is effective in detecting topics in COVID-19 literature, as shown by the evaluation results.
The remainder of this paper is organized as follows. In the next section, we review related works. The Methodology section introduces the structure of the proposed framework, and its system performance is evaluated in the Experiments section. Finally, the conclusions of this research are provided in the Concluding remarks.
Related works
The abovementioned BioNER task aims to recognize biomedical entity boundaries and predict their entity kinds such as genes, proteins, compounds, drugs, mutations and diseases from biomedical literature. However, there are major challenges to accurate identification and classification due to characteristics of biomedical nomenclature such as a lack of standardized naming conventions, frequent crossover in vocabulary, excessive use of abbreviations, synonyms, variants, complex morphology (from the use of unusual characters such as Greek letters), digits, punctuation and many more. Moreover, the biomedical domain is a rapidly evolving field in which new concepts and names are coined on a regular basis. As biomedical concepts are investigated in different disciplines of medicine with distinct naming conventions, new variations are always produced for already existing concepts (8). These new names and concepts make it difficult to extract, classify and comprehend the various formats of terms and often result in the misrecognition of relevant biological entities. Compared with other proper names in generic texts, Biomedical Named Entities (BNEs) pose a greater challenge for existing computer systems.
In response, recent works have adopted advanced NLP technology for BioNER. For instance, Corbett et al. (9) presented word-level and character-level Bi-directional Long Short-Term Memory (BiLSTM) networks for chemical named entity recognition (NER) in the patent literature domain. Hong et al. (10) created a deep learning (DL) architecture, DTranNER, a conditional random fields (CRF)-based framework incorporating a deep learning-based label-label transition model into BioNER, where DL is used to learn the label-label transition relations in an input sequence while considering the context. The DTranNER possesses two distinct DL-based networks: Unary-Network and Pairwise-Network, in which the former is dedicated to individual labeling, while the latter is dedicated to determining the acceptability of label transitions. The CRF of the DL framework is then inputted into these networks. Other models that combine word-level and character-level representations have been utilized in the past. These approaches combine word embeddings with LSTMs (or Bi-LSTMs) over a word’s characters, then pass the representation through another sentence-level Bi-LSTM, and finally predict the final tags using either a softmax or CRF layer.
For biomedical literature classification models, there are two typical categories: bio-entity relation extraction and relevant topic recognition. In the previous decade, the bio-entity relation extraction in the field of Biomedical Natural Language Processing (BioNLP) gained prominence due to the usefulness of identifying key inter-component relationships when summarizing essential knowledge. For instance, protein–protein interaction (PPI) is an important topic in molecular biology because of the growing demand for automatic molecular pathway and interaction discovery from literature. By identifying their participation in the PPI network or comparing them to proteins with similar functionality, it is possible to anticipate the function of uncharacterized proteins. Creating networks of molecular interactions is useful for finding functional modules and discovering new gene-disease correlations. Chang et al. (11) proposed a method that integrates linguistic patterns into a parse tree structure for the support vector machine (SVM) convolution tree kernel to enhance the performance of PPI identification. Another crucial arena in healthcare and other biomedical research is the extraction of chemical–disease relations (CDR) (12).
To encourage exploration, a pioneering challenge of automatically distilling CDRs from the scientific literature was put forward by the BioCreative V organizers (13, 14). This challenge involved identifying chemical-induced disease (CID) linkages from PubMed articles. Prominent methods such as the LSTM network model in conjunction with an SVM model were proposed by Zhou et al. (15). More specifically, LSTM was employed to represent long-range relations in semantics, and the syntactic information was modeled by SVM. A Convolutional Neural Network (CNN) was also proposed by Gu et al. (16) to tackle the CID problem by building a more robust connection representation based on both sequential word order and non-linear dependency pathways, which may naturally reflect the relationships between chemical and illness categories.
The second task of biomedical literature classification is to recognize the relevant topics behind the biomedical text, which can reduce the painstaking challenge of manually curating a huge amount of biomedical literature. This is especially important during the current COVID-19 pandemic, as a large number of clinical, epidemiological and laboratory researches have been conducted to provide policymakers with crucial insights into managing current and future medical and public health issues. This explosive growth of COVID-19 research work has resulted in an increase of around 10 000 research articles per month, with investigation of the disease, its causes and treatments, etc., comprising more than 187 206 articles in PubMed (17). This kind of information overload is a burden among scholars/physicians and can easily hamper efforts in acquiring the latest updates.
A solution to this problem is automated topic prediction, an emerging field in which NLP is used to handle COVID-19-related literature (18). Wahbeh et al. (19), for example, used topic modeling skills to extract important unpublished clinical knowledge from physician social media posts. They uncovered eight subjects, with actions and recommendations being the most prevalent, followed by fighting misinformation. Li et al. (20) used text categorization as well as topic models to quantify temporal variations of stress levels in tweets by users in the USA and identify the sources of stress. A substantial link between stress symptoms and an increase in COVID-19 cases in major US cities was discovered. Moreover, the stress was found to originate and shift from concerns of being infected and other clinical issues to financial worries. Moreover, in 2021, LitCovid (21), an open COVID-19 literature database was developed. More than 100 000 articles have been updated to the database and have reached hundreds of institutions in academia, government and health organizations worldwide with user access of millions. Therefore, the daily maintenance of LitCovid can be burdensome, which includes the labeling of each article to one or more of the predetermined eight related topics, such as Treatment and Diagnosis. This poses a major challenge in the updating process.
Methodology
In this section, we introduce the proposed method for Track 2 (NLM-CHEM) and Track 5 (LitCovid) in the BioCreative VII Challenge. The NLM-CHEM track aims to predict all mentioned chemicals in the full-text article and normalize them to a canonical form. We model it as a sequence labeling problem and define it like this: given a sentence S, which is composed of a sequence of words |$W = \left\{ {{w_1}, {w_2}, \ldots , {w_k}} \right\}$|, for each wr in W, there exists lr in |$L = \left\{ {{l_1}, {l_2}, \ldots , {l_k}} \right\}$| such that each item in W corresponds to its label in L. The purpose of our model is to predict the label of each word in the sequence, and to further identify the entity.
As for the LitCovid track, it involves tackling automated topic annotation for COVID-19 literature, which is a multi-label classification problem and can be formulated as follows. Let |$D = \left\{ {{d_1}, {d_2}, \ldots , {d_n}} \right\}$| be a set of documents, |$T = \left\{ {{t_1}, {t_2}, \ldots , {t_m}} \right\}$| be a set of aspects, where for each topic, there can be one of two possible statuses, |$S = \left\{ {{s_1}, {s_2}} \right\}$| where |${s_1}$|is relevant and |${s_2}$| irrelevant. Thus, for each document |${d_l}$|, and each topic |${t_j}$|, our target is to determine the most suitable state |${s_i}$|. Note that there can be more than one topic for a document.
For both tracks, we propose an ensembled BERT-based approach, as illustrated in Figure 1, that can predict topics and identify bio-entities in the biomedical literature. We first conducted linguistic preprocessing for the input corpus. After that, we adopted multiple pre-trained BERT models to predict topic labels and to identify bio-entities for Track 2 and Track 5, respectively. We further integrate multiple outcomes through an ensemble learning approach for the final output. The following paragraphs describe the design of each layer.
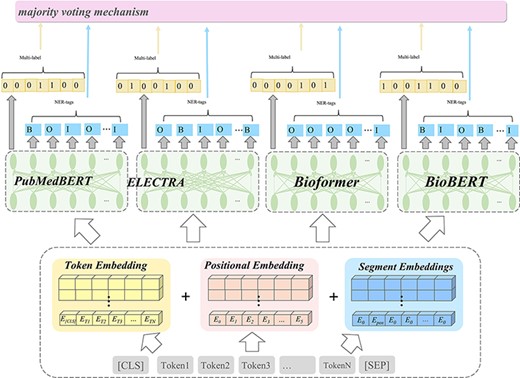
Illustration of the ensemble model in this work for the BioCreative VII challenges.
BERT model for the NLM-CHEM and LitCovid tracks
Input layer—preprocessing and text representation
Preprocessing is crucial in the efficient building of machine learning models. This stage consists of converting all words to lower case characters and removing stop words as well as punctuations. The WordPiece (22) toolkit to represent words by a sequence of smaller tokens is used, and positional embedding tokens (23) were also included. Next, the inputted text sequence is converted to the corresponding sequence format for both tracks respectively.
The NLM-CHEM track facilitates the development of algorithms that can accurately predict chemical entities in biomedical literature and determine which of these chemical entities should be cataloged, and therefore, the track can be considered a NER task. In this track, we need to predict all chemical entities mentioned in the NLM-Chem corpus, in addition to 50 full-text articles that were published in Spring 2021. In light of this, we converted the input text sequence to a labeling sequence. We adopted the BIOE format as the tagging scheme, that is, the word labeled ‘B’ (Begin) and ‘I’ (Inside) means that it is the first and middle or last word of a chemical entity, respectively; the word labeled ‘O’ (Outside) indicates that it does not belong to any chemical entity.
On the other hand, for the LitCovid track, each article can be assigned one or more labels from a set of seven topics (mechanism, transmission, diagnosis, treatment, prevention, case report or epidemic forecasting). Enhancing the accuracy of automated topic prediction in COVID-19-related material is beneficial to researchers worldwide in overcoming information overload. As article titles and abstracts are primarily used to annotate topics, we formulate the topic classification task as a sentence pair classification problem and concatenated the contents of the title and abstract from an article as input text. Finally, the special token ‘[CLS]’ is inserted at the beginning of the sequence, so as to follow the common practice of using pre-trained BERT models for classification tasks.
Multi-head attention layer
Output layer—sequence labeling and multi-label classification
In addition to the pre-trained layers, the last layer of our model consists of a fully connected network with output dimensions of 3 and 7 for NLM-CHEM and LitCovid, respectively. For the output of the NLM-CHEM track, in order to compute the spans in the final evaluation, we take the output of models and rematch them with the original text of the ‘context’ sentence after the output generation has taken place. First, the hidden vectors of the BERT final layer are fed to the output layer with a dropout ratio of 0.3. Then, the Softmax function is applied to the output to obtain a probability distribution for the BIO format labels. The NLM-Chem data has many sub-token entities that are sub-strings of a token rather than the whole string. For example, Gly104Cys has two sub-token entities ‘Gly’ and ‘Cys’. In the objective evaluation, models are supposed to predict the sub-token entities, and not the whole tokens. The majority of sub-token entities occur within mutation names. Approximately 90% of sub-token entities can be treated with simple regular expressions. Consequently, we perform post-processing on sub-token entities, which enhances the performance considerably in the official assessment.
For the LitCovid track, since we considered the LitCovid track to be a multi-label classification problem, we formulated a 7-dim 1D vector, which means that there are seven topics and each topic label is a binary classification of relevant or irrelevant. We used the BCEWithLogitsLoss (https://pytorch.org/docs/stable/generated/torch.nn.BCEWithLogitsLoss.html) as the loss function to alleviate the problem of multi-label. This loss function combines a Sigmoid layer and the binary cross-entropy loss (BCELoss) in one single layer. As a result, it is more stable numerically than a plain Sigmoid layer followed by a BCELoss. By combining the operations into one layer, one takes advantage of the log-sum-exp trick (24) for numerical stability. Applying weighted (In this research, we utilized a widely used heuristic approach which has been included in the scikit-learn package for setting class weight. Please see Appendix A for more detail) BCEWithLogitsLoss can alleviate the problem of data imbalance, and therefore, it has already been popularized in recent research (25–27). More specifically, we employed this loss function to calculate the probability for each topic, and we took the average of losses from all topics as the final loss during model training. The outputs are the topic labels with a class (relevant or irrelevant) and seven possible output states are depicted.
Applying an ensemble learning mechanism to boost model performance
Ensemble learning is accomplished by thresholding the average zero-one decisions of each model per considered label (9). This technique mixes many individual models to improve generalization performance, and the deep learning models with multilayer processing architecture currently outperform shallow or traditional classification models. This inspired us to combine the benefits of both deep learning and ensemble learning and this resulted in a model with improved generalization performance (10). We employed an ensemble learning approach to efficiently solve both tracks. We therefore built a general ensemble learning framework that fuses multiple classifiers created from different pre-trained language models. Since every feature representation is biased and volatile, any single model would be considered a poor classifier in ensemble learning theory (11, 12, 14). Therefore, we trained several different weak classifiers as a group and then combined them for better results. We selected many state-of-the-art pre-trained models as the initialization for the classifier due to consideration of their superior performance in the biomedical domain. By aggregating weak classifiers, the system can effectively minimize the bias and variance of such weak learners, which results in stronger learners with a higher accuracy and more resilient performance.
For the purpose of selecting the most suitable pre-trained model to build our ensemble, we performed 10-fold cross-validation experiments using a variety of BERT models. In the end, we retained BioBERT, PubMedBERT, Sultan and Bioformer for the ensemble. We take the mean of predicted probabilities of each individual classifier and use argmax to obtain the class label. To balance out the individual weaknesses of the five pre-trained BERT models, we also performed more experiments which combined Bioformers, BioBERT, BioM-D, BioM-S and PubmedBERT ensemble models into a single ensemble. We experimented by combining the models into pairs, and all five models using the ensemble method to combine their predictions. The final output of the ensemble was calculated by taking the mean of the predictions from a combination of selected models. Detailed descriptions are shown in Table 1.
| . | Ensembled models for Track . | ||
|---|---|---|---|
| Explanation . | Pre-trained BERT . | NLM-CHEM . | LitCovid . |
| BioBERT: This is the first biomedical-specific BERT model and was proposed by Lee et al. (28). They adopted BERT for the initialized weights and it was pre-trained on large-scale biomedical corpora, PubMed abstracts and PMC full-text articles. It performs well in a variety of biomedical text mining tasks. For the LitCovid track, we use BioBERT v1.2 (https://huggingface.co/dmis-lab/biobert-base-cased-v1.2), which follows the training process of BioBERT v1.1 but includes an LM head, which can be useful for probing. | biobert-base-cased-v1.2 | ✕ | ✓ |
| PubMedBERT: Gu et al. (29) pre-trained this model from scratch using PubMed abstracts with a high batch size (8192), and it showed substantial gains over continual pre-training of general-domain BERT. PubMedBERT achieves state-of-the-art performance on several biomedical NLP tasks, as shown on the Biomedical Language Understanding and Reasoning Benchmark (BLURB) (13). In this research, we adopted PubMedBERT (https://huggingface.co/microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract) for both NLM-CHEM and LitCovid tracks. | PubMedBERT | ✓ | ✓ |
| BioM-Transformers: Alrowili and Shanker (17) pre-trained several large biomedical language models using the original implementation of BERT (30), ALBERT (31) and ELECTRA (18). For both NLM-CHEM and LitCovid tracks, we adopted two kinds of BioM-ELECTRA. One is BioM-ELECTRA-Large-Discriminator (https://huggingface.co/sultan/BioM-ELECTRA-Large-Discriminator), which was pre-trained on PubMed abstracts only with a biomedical domain vocabulary of 434 K steps and a batch size of 4096. The other is BioM-ELECTRA-Large-SQuAD2 (https://huggingface.co/sultan/BioM-ELECTRA-Large-SQuAD2), which fine-tuned BioM-ELECTRA-Large on the SQuAD2.0 dataset. | ELECTRA-Large-Discriminator ELECTRA-Large-SQuAD2 | ✓ ✓ | ✓ ✓ |
| Bioformer: Chen et al. (32) pre-trained Bioformer on all PubMed abstracts (as of Jan 2021) and 1 million randomly-sampled PubMed Central full-text articles. This model achieved the best performance for the LitCovid track in the BioCreative VII Challenge. In this paper, we adopted bioformer-cased-v1.0 (https://huggingface.co/bioformers/bioformer-cased-v1.0) for both NLM-CHEM and LitCovid tracks. In addition, we used bioformer-cased-v1.0-bc2gm (https://huggingface.co/bioformers/bioformer-cased-v1.0-bc2gm), which was fine-tuned on the BC2GM (33) dataset and is suitable for recognizing entities of genes and proteins. | bioformer-cased-v1.0 bioformer-cased-v1.0-bc2gm | ✓ ✓ | ✓ ✕ |
| . | Ensembled models for Track . | ||
|---|---|---|---|
| Explanation . | Pre-trained BERT . | NLM-CHEM . | LitCovid . |
| BioBERT: This is the first biomedical-specific BERT model and was proposed by Lee et al. (28). They adopted BERT for the initialized weights and it was pre-trained on large-scale biomedical corpora, PubMed abstracts and PMC full-text articles. It performs well in a variety of biomedical text mining tasks. For the LitCovid track, we use BioBERT v1.2 (https://huggingface.co/dmis-lab/biobert-base-cased-v1.2), which follows the training process of BioBERT v1.1 but includes an LM head, which can be useful for probing. | biobert-base-cased-v1.2 | ✕ | ✓ |
| PubMedBERT: Gu et al. (29) pre-trained this model from scratch using PubMed abstracts with a high batch size (8192), and it showed substantial gains over continual pre-training of general-domain BERT. PubMedBERT achieves state-of-the-art performance on several biomedical NLP tasks, as shown on the Biomedical Language Understanding and Reasoning Benchmark (BLURB) (13). In this research, we adopted PubMedBERT (https://huggingface.co/microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract) for both NLM-CHEM and LitCovid tracks. | PubMedBERT | ✓ | ✓ |
| BioM-Transformers: Alrowili and Shanker (17) pre-trained several large biomedical language models using the original implementation of BERT (30), ALBERT (31) and ELECTRA (18). For both NLM-CHEM and LitCovid tracks, we adopted two kinds of BioM-ELECTRA. One is BioM-ELECTRA-Large-Discriminator (https://huggingface.co/sultan/BioM-ELECTRA-Large-Discriminator), which was pre-trained on PubMed abstracts only with a biomedical domain vocabulary of 434 K steps and a batch size of 4096. The other is BioM-ELECTRA-Large-SQuAD2 (https://huggingface.co/sultan/BioM-ELECTRA-Large-SQuAD2), which fine-tuned BioM-ELECTRA-Large on the SQuAD2.0 dataset. | ELECTRA-Large-Discriminator ELECTRA-Large-SQuAD2 | ✓ ✓ | ✓ ✓ |
| Bioformer: Chen et al. (32) pre-trained Bioformer on all PubMed abstracts (as of Jan 2021) and 1 million randomly-sampled PubMed Central full-text articles. This model achieved the best performance for the LitCovid track in the BioCreative VII Challenge. In this paper, we adopted bioformer-cased-v1.0 (https://huggingface.co/bioformers/bioformer-cased-v1.0) for both NLM-CHEM and LitCovid tracks. In addition, we used bioformer-cased-v1.0-bc2gm (https://huggingface.co/bioformers/bioformer-cased-v1.0-bc2gm), which was fine-tuned on the BC2GM (33) dataset and is suitable for recognizing entities of genes and proteins. | bioformer-cased-v1.0 bioformer-cased-v1.0-bc2gm | ✓ ✓ | ✓ ✕ |
| . | Ensembled models for Track . | ||
|---|---|---|---|
| Explanation . | Pre-trained BERT . | NLM-CHEM . | LitCovid . |
| BioBERT: This is the first biomedical-specific BERT model and was proposed by Lee et al. (28). They adopted BERT for the initialized weights and it was pre-trained on large-scale biomedical corpora, PubMed abstracts and PMC full-text articles. It performs well in a variety of biomedical text mining tasks. For the LitCovid track, we use BioBERT v1.2 (https://huggingface.co/dmis-lab/biobert-base-cased-v1.2), which follows the training process of BioBERT v1.1 but includes an LM head, which can be useful for probing. | biobert-base-cased-v1.2 | ✕ | ✓ |
| PubMedBERT: Gu et al. (29) pre-trained this model from scratch using PubMed abstracts with a high batch size (8192), and it showed substantial gains over continual pre-training of general-domain BERT. PubMedBERT achieves state-of-the-art performance on several biomedical NLP tasks, as shown on the Biomedical Language Understanding and Reasoning Benchmark (BLURB) (13). In this research, we adopted PubMedBERT (https://huggingface.co/microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract) for both NLM-CHEM and LitCovid tracks. | PubMedBERT | ✓ | ✓ |
| BioM-Transformers: Alrowili and Shanker (17) pre-trained several large biomedical language models using the original implementation of BERT (30), ALBERT (31) and ELECTRA (18). For both NLM-CHEM and LitCovid tracks, we adopted two kinds of BioM-ELECTRA. One is BioM-ELECTRA-Large-Discriminator (https://huggingface.co/sultan/BioM-ELECTRA-Large-Discriminator), which was pre-trained on PubMed abstracts only with a biomedical domain vocabulary of 434 K steps and a batch size of 4096. The other is BioM-ELECTRA-Large-SQuAD2 (https://huggingface.co/sultan/BioM-ELECTRA-Large-SQuAD2), which fine-tuned BioM-ELECTRA-Large on the SQuAD2.0 dataset. | ELECTRA-Large-Discriminator ELECTRA-Large-SQuAD2 | ✓ ✓ | ✓ ✓ |
| Bioformer: Chen et al. (32) pre-trained Bioformer on all PubMed abstracts (as of Jan 2021) and 1 million randomly-sampled PubMed Central full-text articles. This model achieved the best performance for the LitCovid track in the BioCreative VII Challenge. In this paper, we adopted bioformer-cased-v1.0 (https://huggingface.co/bioformers/bioformer-cased-v1.0) for both NLM-CHEM and LitCovid tracks. In addition, we used bioformer-cased-v1.0-bc2gm (https://huggingface.co/bioformers/bioformer-cased-v1.0-bc2gm), which was fine-tuned on the BC2GM (33) dataset and is suitable for recognizing entities of genes and proteins. | bioformer-cased-v1.0 bioformer-cased-v1.0-bc2gm | ✓ ✓ | ✓ ✕ |
| . | Ensembled models for Track . | ||
|---|---|---|---|
| Explanation . | Pre-trained BERT . | NLM-CHEM . | LitCovid . |
| BioBERT: This is the first biomedical-specific BERT model and was proposed by Lee et al. (28). They adopted BERT for the initialized weights and it was pre-trained on large-scale biomedical corpora, PubMed abstracts and PMC full-text articles. It performs well in a variety of biomedical text mining tasks. For the LitCovid track, we use BioBERT v1.2 (https://huggingface.co/dmis-lab/biobert-base-cased-v1.2), which follows the training process of BioBERT v1.1 but includes an LM head, which can be useful for probing. | biobert-base-cased-v1.2 | ✕ | ✓ |
| PubMedBERT: Gu et al. (29) pre-trained this model from scratch using PubMed abstracts with a high batch size (8192), and it showed substantial gains over continual pre-training of general-domain BERT. PubMedBERT achieves state-of-the-art performance on several biomedical NLP tasks, as shown on the Biomedical Language Understanding and Reasoning Benchmark (BLURB) (13). In this research, we adopted PubMedBERT (https://huggingface.co/microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract) for both NLM-CHEM and LitCovid tracks. | PubMedBERT | ✓ | ✓ |
| BioM-Transformers: Alrowili and Shanker (17) pre-trained several large biomedical language models using the original implementation of BERT (30), ALBERT (31) and ELECTRA (18). For both NLM-CHEM and LitCovid tracks, we adopted two kinds of BioM-ELECTRA. One is BioM-ELECTRA-Large-Discriminator (https://huggingface.co/sultan/BioM-ELECTRA-Large-Discriminator), which was pre-trained on PubMed abstracts only with a biomedical domain vocabulary of 434 K steps and a batch size of 4096. The other is BioM-ELECTRA-Large-SQuAD2 (https://huggingface.co/sultan/BioM-ELECTRA-Large-SQuAD2), which fine-tuned BioM-ELECTRA-Large on the SQuAD2.0 dataset. | ELECTRA-Large-Discriminator ELECTRA-Large-SQuAD2 | ✓ ✓ | ✓ ✓ |
| Bioformer: Chen et al. (32) pre-trained Bioformer on all PubMed abstracts (as of Jan 2021) and 1 million randomly-sampled PubMed Central full-text articles. This model achieved the best performance for the LitCovid track in the BioCreative VII Challenge. In this paper, we adopted bioformer-cased-v1.0 (https://huggingface.co/bioformers/bioformer-cased-v1.0) for both NLM-CHEM and LitCovid tracks. In addition, we used bioformer-cased-v1.0-bc2gm (https://huggingface.co/bioformers/bioformer-cased-v1.0-bc2gm), which was fine-tuned on the BC2GM (33) dataset and is suitable for recognizing entities of genes and proteins. | bioformer-cased-v1.0 bioformer-cased-v1.0-bc2gm | ✓ ✓ | ✓ ✕ |
| MeSH ID Normalization Algorithm |
| INPUT: E = {e1,, ec}—a set of all predicted chemical named entities; K = {k1:v1,, km:vm}—a set of key-value pairs in the MeSH ID dictionary |
| BEGIN |
| 1: FOR EACH PREDICTED ENTITY er: |
| 2: FOR i = 1 TO m |
| 3: IF er == ki |
| 4: RETURN vi AS MeSH ID |
| 5: ELSE |
| 6: calculate dist = Levenshtein Distance(er, ki) |
| 7: IF dist ≥ 90 |
| 8: RETURN vi AS MeSH ID |
| 9: ELSE |
| 10: RETURN ‘-’ AS empty value |
| 11: END FOR |
| 12: END FOR EACH PREDICTED ENTITY |
| END |
| MeSH ID Normalization Algorithm |
| INPUT: E = {e1,, ec}—a set of all predicted chemical named entities; K = {k1:v1,, km:vm}—a set of key-value pairs in the MeSH ID dictionary |
| BEGIN |
| 1: FOR EACH PREDICTED ENTITY er: |
| 2: FOR i = 1 TO m |
| 3: IF er == ki |
| 4: RETURN vi AS MeSH ID |
| 5: ELSE |
| 6: calculate dist = Levenshtein Distance(er, ki) |
| 7: IF dist ≥ 90 |
| 8: RETURN vi AS MeSH ID |
| 9: ELSE |
| 10: RETURN ‘-’ AS empty value |
| 11: END FOR |
| 12: END FOR EACH PREDICTED ENTITY |
| END |
| MeSH ID Normalization Algorithm |
| INPUT: E = {e1,, ec}—a set of all predicted chemical named entities; K = {k1:v1,, km:vm}—a set of key-value pairs in the MeSH ID dictionary |
| BEGIN |
| 1: FOR EACH PREDICTED ENTITY er: |
| 2: FOR i = 1 TO m |
| 3: IF er == ki |
| 4: RETURN vi AS MeSH ID |
| 5: ELSE |
| 6: calculate dist = Levenshtein Distance(er, ki) |
| 7: IF dist ≥ 90 |
| 8: RETURN vi AS MeSH ID |
| 9: ELSE |
| 10: RETURN ‘-’ AS empty value |
| 11: END FOR |
| 12: END FOR EACH PREDICTED ENTITY |
| END |
| MeSH ID Normalization Algorithm |
| INPUT: E = {e1,, ec}—a set of all predicted chemical named entities; K = {k1:v1,, km:vm}—a set of key-value pairs in the MeSH ID dictionary |
| BEGIN |
| 1: FOR EACH PREDICTED ENTITY er: |
| 2: FOR i = 1 TO m |
| 3: IF er == ki |
| 4: RETURN vi AS MeSH ID |
| 5: ELSE |
| 6: calculate dist = Levenshtein Distance(er, ki) |
| 7: IF dist ≥ 90 |
| 8: RETURN vi AS MeSH ID |
| 9: ELSE |
| 10: RETURN ‘-’ AS empty value |
| 11: END FOR |
| 12: END FOR EACH PREDICTED ENTITY |
| END |
An edit distance-based entity linking approach for chemical name normalization
In this research, we employ the edit distance algorithm to address the entity linking problem in chemical normalization. At the outset, a collection of MeSH ID and identifications from the dataset were compiled into a knowledge base (dictionary). During prediction, we search for predicted chemical named entities in the dictionary in order to find the correct mapping. In the case of missing entities, we calculated the Levenshtein Distance (34) with a 90% similarity level to obtain the most similar terms and their ID. Finally, if the above process yields no return, we designate the term with a null value. Practically, we implement this with thefuzz (https://github.com/seatgeek/thefuzz) and python-Levenshtein (https://github.com/ztane/python-Levenshtein/) python packages. The chemical named entity normalization algorithm is presented as follows:
There are a set of predicted chemical named entities (|$E$|) to map a set of key-value pairs in the MeSH ID (|$K$|) over several commits (|$n$|), and a total of |$\left( {E \cdot K} \right) \times n$| processes are performed. We employed parallel threading to speed up the search process. The total search time was reduced to within an hour using 20 CPU cores. The current framework was shown to achieve remarkable performance, surpassing baseline as well as the median of all compared methods.
Experiments
Dataset & setting
The NLM-CHEM track uses the NLM-CHEM corpus (35) for the training and development sets. This corpus includes 150 full-text articles with about 5000 unique chemical named entities that are mapped to approximately 2000 MeSH identifiers. The test set is a collection of 50 recently published full-text articles on PubMed, planned to be indexed manually in the year 2021. More specifically, there are 3740 unique chemical strings and 1352 unique MeSH IDs in the test set. The average number of Chemical Annotations per article is 300.4 terms, but there is a minimum of 2 terms and a maximum of 1318 terms The distribution of the number of unique MeSH IDs per article is also similar, with a minimum of 1 and an average of 41, but the largest number of unique MeSH IDs in an article is 127.
The LitCovid track employs the LitCovid corpus (21) for multi-label topic classification of the COVID-19 literature. The training and development sets contain more than 30 000 COVID-19 related articles and the evaluation dataset includes 2500 manually reviewed articles. The abstract and title of an article along with other meta-information, such as DOI, journal name and keywords, may contain one or more labels. The labels include Treatment, Diagnosis, Prevention, Mechanisms, Transmission, Epidemiological Prediction and Case Reporting. Detailed information of the corpora used in both tracks is listed in Table 2.
The data distribution of the NLM-CHEM and LitCovid tracks in the BioCreative VII Challenge
| . | Training . | Development . | Test . |
|---|---|---|---|
| NLM-CHEM200 Corpus | |||
| # of Articles | 100 | 50 | 54 |
| # of Chemical NE (those with a MeSH ID) | 26 567 (26 339) | 11 772 (11 660) | 22 942 (22 777) |
| LitCovid Corpus | |||
| # of Articles | 24 960 | 6239 | 2500 |
| # of Prevention | 11 102 (44.48%) | 2750 (44.08%) | 1035 (41.4%) |
| # of Treatment | 8717 (34.2%) | 2207 (35.37%) | 722 (28.88%) |
| # of Diagnosis | 6193 (24.81%) | 1546 (24.78%) | 926 (37.04%) |
| # of Mechanism | 4438 (17.78%) | 1073 (17.2%) | 567 (22.68%) |
| # of Case report | 2063 (8.27%) | 482 (7.72%) | 128 (5.12%) |
| # of Transmission | 1088 (4.35%) | 256 (4.1%) | 41 (1.64%) |
| # of Epidemic forecasting | 645 (2.58%) | 192 (3.08%) | 197 (7.88%) |
| . | Training . | Development . | Test . |
|---|---|---|---|
| NLM-CHEM200 Corpus | |||
| # of Articles | 100 | 50 | 54 |
| # of Chemical NE (those with a MeSH ID) | 26 567 (26 339) | 11 772 (11 660) | 22 942 (22 777) |
| LitCovid Corpus | |||
| # of Articles | 24 960 | 6239 | 2500 |
| # of Prevention | 11 102 (44.48%) | 2750 (44.08%) | 1035 (41.4%) |
| # of Treatment | 8717 (34.2%) | 2207 (35.37%) | 722 (28.88%) |
| # of Diagnosis | 6193 (24.81%) | 1546 (24.78%) | 926 (37.04%) |
| # of Mechanism | 4438 (17.78%) | 1073 (17.2%) | 567 (22.68%) |
| # of Case report | 2063 (8.27%) | 482 (7.72%) | 128 (5.12%) |
| # of Transmission | 1088 (4.35%) | 256 (4.1%) | 41 (1.64%) |
| # of Epidemic forecasting | 645 (2.58%) | 192 (3.08%) | 197 (7.88%) |
The data distribution of the NLM-CHEM and LitCovid tracks in the BioCreative VII Challenge
| . | Training . | Development . | Test . |
|---|---|---|---|
| NLM-CHEM200 Corpus | |||
| # of Articles | 100 | 50 | 54 |
| # of Chemical NE (those with a MeSH ID) | 26 567 (26 339) | 11 772 (11 660) | 22 942 (22 777) |
| LitCovid Corpus | |||
| # of Articles | 24 960 | 6239 | 2500 |
| # of Prevention | 11 102 (44.48%) | 2750 (44.08%) | 1035 (41.4%) |
| # of Treatment | 8717 (34.2%) | 2207 (35.37%) | 722 (28.88%) |
| # of Diagnosis | 6193 (24.81%) | 1546 (24.78%) | 926 (37.04%) |
| # of Mechanism | 4438 (17.78%) | 1073 (17.2%) | 567 (22.68%) |
| # of Case report | 2063 (8.27%) | 482 (7.72%) | 128 (5.12%) |
| # of Transmission | 1088 (4.35%) | 256 (4.1%) | 41 (1.64%) |
| # of Epidemic forecasting | 645 (2.58%) | 192 (3.08%) | 197 (7.88%) |
| . | Training . | Development . | Test . |
|---|---|---|---|
| NLM-CHEM200 Corpus | |||
| # of Articles | 100 | 50 | 54 |
| # of Chemical NE (those with a MeSH ID) | 26 567 (26 339) | 11 772 (11 660) | 22 942 (22 777) |
| LitCovid Corpus | |||
| # of Articles | 24 960 | 6239 | 2500 |
| # of Prevention | 11 102 (44.48%) | 2750 (44.08%) | 1035 (41.4%) |
| # of Treatment | 8717 (34.2%) | 2207 (35.37%) | 722 (28.88%) |
| # of Diagnosis | 6193 (24.81%) | 1546 (24.78%) | 926 (37.04%) |
| # of Mechanism | 4438 (17.78%) | 1073 (17.2%) | 567 (22.68%) |
| # of Case report | 2063 (8.27%) | 482 (7.72%) | 128 (5.12%) |
| # of Transmission | 1088 (4.35%) | 256 (4.1%) | 41 (1.64%) |
| # of Epidemic forecasting | 645 (2.58%) | 192 (3.08%) | 197 (7.88%) |
The metrics used to evaluate the prediction performance of the NLM-CHEM track are precision, recall and F1-score (in ‘strict’ and ‘approximate’ evaluation settings), as well as the micro-average used for comparing the overall performance. Specifically, for both NER and normalization tasks, the ‘strict’ setting expects an exact match between two spans, i.e. the predicted span of an entity/MeSH ID and the correctly annotated span/ID. On the other hand, the ‘approximate’ metric for the NER task considers a span as correct if it overlaps with the gold span.
As for the LitCovid track, the two most widely utilized metrics for multi-label categorization are label-based and instance-based assessment measures (36). Label-based evaluation independently judges each label, with associated measures calculating each label’s performance before aggregating the results for all labels. Instance-based measures, on the other hand, treat every instance as a separate entity. Similar to the NLM-CHEM track, this track also evaluates the precision, recall and F1-score of instance-based results. The macro- and micro-averages were further adopted for estimating the performance of label-based matching.
The proposed model was implemented using PyTorch (https://pytorch.org/), a Python deep learning library. We adopted the common settings of optimizer and hyper-parameters for fine-tuning, i.e. 10 epochs of training time with the AdamW optimizer (37) using a learning rate of 2e-5. However, the weight decay was set to 1e-3 to improve stability during training. The batch size of 16 and 64 were set for the NLM-CHEM and LitCovid tracks, respectively. The maximum sequence length was 512 tokens, with padding or truncating at the end of the sequence. We ran the proposed model on two NVIDIA GeForce RTX 3090 GPUs.
Results and discussion
To conduct a comprehensive evaluation, we listed the benchmarks (BlueBERT (38) for NLM-CHEM; ML-Net (39) for LitCovid), median performance of the participating teams (MPT), and the top one system (T1S (40) for NLM-CHEM; Bioformer (41) for LitCovid), from both tracks as comparisons. Moreover, we also selected a collection of BERT variants that were used in our ensemble learning approach: BioBERT, PubMedBERT, BioM-ELECTRA-Large-Discriminator (BioM-D), BioM-ELECTRA-Large-SQuAD2 (BioM-S), bioformer-cased-v1.0 (Bioformer) for both tracks; and bioformer-cased-v1.0-bc2gm (Bioformer-B) for the LitCovid track, as comparisons.
Table 3 presents the performance of the Bioformer and the results of incrementally applying different pre-trained BERT models in the NLM-CHEM track. The performance can be further improved by integrating different BERT models incrementally under the ensemble learning framework. Consequently, applying them altogether achieves the best performance. In addition, we investigated the impact of different data sizes as shown in Table 4. In general, our system performance is not significantly affected by data size. The impact is relatively large only when there is only 10% of the data, in which the performance is greatly reduced with more than 10% reduction in the F1-scores. The results showed that the proposed method is robust and efficient in both tracks.
Incremental contribution of different BERT models for ensemble learning in the NLM-CHEM track
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| +Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7162/0.8635/0.7796 |
| +BioM-D | 0.8299/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7707/0.8312/0.7988 | 0.7271/0.8616/0.7856 |
| +BioM-S | 0.8294/0.8627/0.8457 | 0.8969/0.9276/0.9120 | 0.7697/0.8303/0.7988 | 0.7247/0.8588/0.7826 |
| +PubMedBERT | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| +Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7162/0.8635/0.7796 |
| +BioM-D | 0.8299/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7707/0.8312/0.7988 | 0.7271/0.8616/0.7856 |
| +BioM-S | 0.8294/0.8627/0.8457 | 0.8969/0.9276/0.9120 | 0.7697/0.8303/0.7988 | 0.7247/0.8588/0.7826 |
| +PubMedBERT | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
Incremental contribution of different BERT models for ensemble learning in the NLM-CHEM track
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| +Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7162/0.8635/0.7796 |
| +BioM-D | 0.8299/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7707/0.8312/0.7988 | 0.7271/0.8616/0.7856 |
| +BioM-S | 0.8294/0.8627/0.8457 | 0.8969/0.9276/0.9120 | 0.7697/0.8303/0.7988 | 0.7247/0.8588/0.7826 |
| +PubMedBERT | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| +Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7162/0.8635/0.7796 |
| +BioM-D | 0.8299/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7707/0.8312/0.7988 | 0.7271/0.8616/0.7856 |
| +BioM-S | 0.8294/0.8627/0.8457 | 0.8969/0.9276/0.9120 | 0.7697/0.8303/0.7988 | 0.7247/0.8588/0.7826 |
| +PubMedBERT | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Data size . | Precision/Recall/F1-score . | |||
| 10% | 0.7029/0.7247/0.7136 | 0.8296/0.8239/0.8268 | 0.6726/0.7414/0.7053 | 0.6425/0.7957/0.7061 |
| 20% | 0.7679/0.8355/0.8003 | 0.8498/0.9138/0.8806 | 0.7276/0.8045/0.7641 | 0.6782/0.8438/0.7487 |
| 50% | 0.8018/0.8680/0.8336 | 0.8776/0.9420/0.9087 | 0.7494/0.8257/0.7857 | 0.7060/0.8583/0.7704 |
| 100% | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Data size . | Precision/Recall/F1-score . | |||
| 10% | 0.7029/0.7247/0.7136 | 0.8296/0.8239/0.8268 | 0.6726/0.7414/0.7053 | 0.6425/0.7957/0.7061 |
| 20% | 0.7679/0.8355/0.8003 | 0.8498/0.9138/0.8806 | 0.7276/0.8045/0.7641 | 0.6782/0.8438/0.7487 |
| 50% | 0.8018/0.8680/0.8336 | 0.8776/0.9420/0.9087 | 0.7494/0.8257/0.7857 | 0.7060/0.8583/0.7704 |
| 100% | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Data size . | Precision/Recall/F1-score . | |||
| 10% | 0.7029/0.7247/0.7136 | 0.8296/0.8239/0.8268 | 0.6726/0.7414/0.7053 | 0.6425/0.7957/0.7061 |
| 20% | 0.7679/0.8355/0.8003 | 0.8498/0.9138/0.8806 | 0.7276/0.8045/0.7641 | 0.6782/0.8438/0.7487 |
| 50% | 0.8018/0.8680/0.8336 | 0.8776/0.9420/0.9087 | 0.7494/0.8257/0.7857 | 0.7060/0.8583/0.7704 |
| 100% | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Data size . | Precision/Recall/F1-score . | |||
| 10% | 0.7029/0.7247/0.7136 | 0.8296/0.8239/0.8268 | 0.6726/0.7414/0.7053 | 0.6425/0.7957/0.7061 |
| 20% | 0.7679/0.8355/0.8003 | 0.8498/0.9138/0.8806 | 0.7276/0.8045/0.7641 | 0.6782/0.8438/0.7487 |
| 50% | 0.8018/0.8680/0.8336 | 0.8776/0.9420/0.9087 | 0.7494/0.8257/0.7857 | 0.7060/0.8583/0.7704 |
| 100% | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
Table 5 presents the performance comparisons in the NLM-CHEM track. The overall outcome of BioBERT is F1-scores of about 80% and 86% in strict and approximate evaluations, respectively, which is generally worse than all of the compared methods. This is most likely due to the fact that it is the first biomedical-specific BERT, and the scale of the training dataset is smaller than the other compared systems In contrast, BlueBERT pre-trained on the BLUE (Biomedical Language Understanding Evaluation) dataset (38), a much more complex corpus consisting of five tasks with ten datasets that covered both biomedical and clinical articles of various sizes and challenges. Hence, it surpassed BioBERT by about 2% in terms of F1-score. Furthermore, the PubMedBERT, BioM and Bioformer employed more pre-training data, and therefore, achieved a more fine-tuned performance with F1-scores of 83% and 90% in the strict and approximate evaluations, respectively. Their performances significantly outperformed the BlueBERT, and they were even superior to the median performance of the participating teams in this track. It is noteworthy that the ensemble learning-based method, T1S, and our proposed method can further enhance the overall performances by 3%, therefore achieving F1-scores of 86% and 92% in the strict and approximate evaluations, respectively. This indicates that integrating multiple BERT models can advance the performance for full-text chemical identification significantly. It is interesting to note that T1S achieved the best precision, and the reason for this is that the tagging consistency and entity coverage are improved through majority voting. The ensemble method of T1S focused on the inconsistent predictions in the same article, and it computed the majority for model predictions and changed all the minority predictions to the majority label. In this way, the ensemble mechanism was the majority voting from all predictions from individual models within an article. Our method thus achieved the best recall. We postulate that because our ensemble approach integrated multiple outputs from different BERT models, it obtained a better generalization of the textual structures of chemical entities. This, therefore, facilitated the learning of characteristics of chemical identification for each structural type, which in turn increased the recall rate. In addition, our proposed MeSH ID normalization algorithm is effective in chemical entity normalization, which achieved F1-score of about 80% in both strict and approximate evaluations. It is observed that the strict and approximate scores do not differ much. This is possibly due to the short token length, which resulted in the efficient use of the edit distance-based method to partially match token sequences in search of the correct answer in the MeSH hierarchy.
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| BlueBERT | 0.8440/0.7877/0.8149 | 0.9156/0.8492/0.8811 | 0.8151/0.7644/0.7899 | 0.7917/0.7889/0.7857 |
| MPT | 0.8476/0.8136/0.8373 | 0.9220/0.8682/0.8951 | 0.7120/0.7760/0.7749 | 0.6782/0.8402/0.7552 |
| BioBERT | 0.8010/0.7830/0.7919 | 0.8773/0.8528/0.8649 | 0.7582/0.8205/0.7881 | 0.7096/0.8497/0.7690 |
| PubMedBERT | 0.8488/0.8542/0.8515 | 0.9184/0.9171/0.9177 | 0.7788/0.8272/0.8023 | 0.7354/0.8586/0.7889 |
| BioM-S | 0.8583/0.8457/0.8520 | 0.9246/0.9055/0.9149 | 0.7816/0.8290/0.8046 | 0.7374/0.8613/0.7898 |
| BioM-D | 0.8520/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7840/0.8275/0.8052 | 0.7432/0.8566/0.7923 |
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7166/0.8626/0.7793 |
| T1S | 0.8759/0.8587/0.8672 | 0.9373/0.9161/0.9266 | 0.8621/0.7702/0.8136 | 0.8302/0.7867/0.8030 |
| Our method | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| BlueBERT | 0.8440/0.7877/0.8149 | 0.9156/0.8492/0.8811 | 0.8151/0.7644/0.7899 | 0.7917/0.7889/0.7857 |
| MPT | 0.8476/0.8136/0.8373 | 0.9220/0.8682/0.8951 | 0.7120/0.7760/0.7749 | 0.6782/0.8402/0.7552 |
| BioBERT | 0.8010/0.7830/0.7919 | 0.8773/0.8528/0.8649 | 0.7582/0.8205/0.7881 | 0.7096/0.8497/0.7690 |
| PubMedBERT | 0.8488/0.8542/0.8515 | 0.9184/0.9171/0.9177 | 0.7788/0.8272/0.8023 | 0.7354/0.8586/0.7889 |
| BioM-S | 0.8583/0.8457/0.8520 | 0.9246/0.9055/0.9149 | 0.7816/0.8290/0.8046 | 0.7374/0.8613/0.7898 |
| BioM-D | 0.8520/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7840/0.8275/0.8052 | 0.7432/0.8566/0.7923 |
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7166/0.8626/0.7793 |
| T1S | 0.8759/0.8587/0.8672 | 0.9373/0.9161/0.9266 | 0.8621/0.7702/0.8136 | 0.8302/0.7867/0.8030 |
| Our method | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| BlueBERT | 0.8440/0.7877/0.8149 | 0.9156/0.8492/0.8811 | 0.8151/0.7644/0.7899 | 0.7917/0.7889/0.7857 |
| MPT | 0.8476/0.8136/0.8373 | 0.9220/0.8682/0.8951 | 0.7120/0.7760/0.7749 | 0.6782/0.8402/0.7552 |
| BioBERT | 0.8010/0.7830/0.7919 | 0.8773/0.8528/0.8649 | 0.7582/0.8205/0.7881 | 0.7096/0.8497/0.7690 |
| PubMedBERT | 0.8488/0.8542/0.8515 | 0.9184/0.9171/0.9177 | 0.7788/0.8272/0.8023 | 0.7354/0.8586/0.7889 |
| BioM-S | 0.8583/0.8457/0.8520 | 0.9246/0.9055/0.9149 | 0.7816/0.8290/0.8046 | 0.7374/0.8613/0.7898 |
| BioM-D | 0.8520/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7840/0.8275/0.8052 | 0.7432/0.8566/0.7923 |
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7166/0.8626/0.7793 |
| T1S | 0.8759/0.8587/0.8672 | 0.9373/0.9161/0.9266 | 0.8621/0.7702/0.8136 | 0.8302/0.7867/0.8030 |
| Our method | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
| . | Chemical mention recognition . | Chemical normalization to MeSH IDs . | ||
|---|---|---|---|---|
| . | Strict . | Approximate . | Strict . | Approximate . |
| Systems . | Precision/Recall/F1-score . | |||
| BlueBERT | 0.8440/0.7877/0.8149 | 0.9156/0.8492/0.8811 | 0.8151/0.7644/0.7899 | 0.7917/0.7889/0.7857 |
| MPT | 0.8476/0.8136/0.8373 | 0.9220/0.8682/0.8951 | 0.7120/0.7760/0.7749 | 0.6782/0.8402/0.7552 |
| BioBERT | 0.8010/0.7830/0.7919 | 0.8773/0.8528/0.8649 | 0.7582/0.8205/0.7881 | 0.7096/0.8497/0.7690 |
| PubMedBERT | 0.8488/0.8542/0.8515 | 0.9184/0.9171/0.9177 | 0.7788/0.8272/0.8023 | 0.7354/0.8586/0.7889 |
| BioM-S | 0.8583/0.8457/0.8520 | 0.9246/0.9055/0.9149 | 0.7816/0.8290/0.8046 | 0.7374/0.8613/0.7898 |
| BioM-D | 0.8520/0.8419/0.8469 | 0.9216/0.9052/0.9133 | 0.7840/0.8275/0.8052 | 0.7432/0.8566/0.7923 |
| Bioformer | 0.8156/0.8576/0.8361 | 0.8846/0.9236/0.9037 | 0.7570/0.8294/0.7915 | 0.7130/0.8596/0.7756 |
| Bioformer-B | 0.8140/0.8558/0.8344 | 0.8847/0.9249/0.9044 | 0.7652/0.8306/0.7965 | 0.7166/0.8626/0.7793 |
| T1S | 0.8759/0.8587/0.8672 | 0.9373/0.9161/0.9266 | 0.8621/0.7702/0.8136 | 0.8302/0.7867/0.8030 |
| Our method | 0.8535/0.8622/0.8578 | 0.9201/0.9237/0.9219 | 0.7835/0.8303/0.8062 | 0.7448/0.8570/0.7933 |
For the performance evaluation of the LitCovid track, Table 6 illustrates the incremental performance of utilizing different pre-trained BERT models, and Table 7 presents the impact on performance due to different data sizes. The results are identical to the NLM-CHEM track, in which integrating effective models altogether achieved the best performance, and the proposed BERT-based ensemble approach is not only efficient but also robust due to its ability to achieve remarkable performance with different sizes of the dataset. Table 8 displays the performances of the compared systems on the multi-label topic classification in the LitCovid track. The baseline method, ML-Net, is a BiLSTM-based neural network. It had a mediocre performance with F1-scores of 76.6% and 86.8 on the label-based macro average and instance-based, respectively. The BERT-based models can significantly improve the performance by about 10% in F1-score in both label-based and instance-based evaluations. Interestingly, BioBERT outperformed almost all of the comparisons and is comparable to Bioformer, which differs from the performance obtained in the NLM-CHEM track. The Bioformer was pre-trained on the three different sources of abstracts from PubMed, full-text from one million PMC articles, and approximately 20 000 abstracts of COVID-19 publications. It thus achieved the best performance among the participating teams in the LitCovid track. In this paper, we used the Bioformer to integrate the advantages of different BERT models by means of a majority voting mechanism. For this reason, the proposed method outperformed all of the comparisons and achieved the state-of-the-art performance on the LitCovid corpus.
Incremental contribution of different BERT models for ensemble learning in the LitCovid track
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| +BioBERT | 0.9170/0.9165/0.9167 | 0.8815/0.8902/0.8818 | 0.9355/0.9367/0.9361 |
| +BioM-S | 0.9240/0.9140/0.9189 | 0.9001/0.8759/0.8858 | 0.9403/0.9357/0.9380 |
| +PubMedBERT | 0.9303/0.9076/0.9188 | 0.9128/0.8681/0.8865 | 0.9454/0.9321/0.9387 |
| +BioM-D | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| +BioBERT | 0.9170/0.9165/0.9167 | 0.8815/0.8902/0.8818 | 0.9355/0.9367/0.9361 |
| +BioM-S | 0.9240/0.9140/0.9189 | 0.9001/0.8759/0.8858 | 0.9403/0.9357/0.9380 |
| +PubMedBERT | 0.9303/0.9076/0.9188 | 0.9128/0.8681/0.8865 | 0.9454/0.9321/0.9387 |
| +BioM-D | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
Incremental contribution of different BERT models for ensemble learning in the LitCovid track
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| +BioBERT | 0.9170/0.9165/0.9167 | 0.8815/0.8902/0.8818 | 0.9355/0.9367/0.9361 |
| +BioM-S | 0.9240/0.9140/0.9189 | 0.9001/0.8759/0.8858 | 0.9403/0.9357/0.9380 |
| +PubMedBERT | 0.9303/0.9076/0.9188 | 0.9128/0.8681/0.8865 | 0.9454/0.9321/0.9387 |
| +BioM-D | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| +BioBERT | 0.9170/0.9165/0.9167 | 0.8815/0.8902/0.8818 | 0.9355/0.9367/0.9361 |
| +BioM-S | 0.9240/0.9140/0.9189 | 0.9001/0.8759/0.8858 | 0.9403/0.9357/0.9380 |
| +PubMedBERT | 0.9303/0.9076/0.9188 | 0.9128/0.8681/0.8865 | 0.9454/0.9321/0.9387 |
| +BioM-D | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Data size . | Precision/Recall/F1-score . | ||
| 10% | 0.9169/0.8908/0.9036 | 0.9087/0.8230/0.8537 | 0.9308/0.9179/0.9243 |
| 20% | 0.9242/0.9002/0.9120 | 0.9089/0.8455/0.8690 | 0.9387/0.9255/0.9321 |
| 50% | 0.9250/0.9137/0.9193 | 0.9114/0.8642/0.8826 | 0.9419/0.9354/0.9386 |
| 100% | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Data size . | Precision/Recall/F1-score . | ||
| 10% | 0.9169/0.8908/0.9036 | 0.9087/0.8230/0.8537 | 0.9308/0.9179/0.9243 |
| 20% | 0.9242/0.9002/0.9120 | 0.9089/0.8455/0.8690 | 0.9387/0.9255/0.9321 |
| 50% | 0.9250/0.9137/0.9193 | 0.9114/0.8642/0.8826 | 0.9419/0.9354/0.9386 |
| 100% | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Data size . | Precision/Recall/F1-score . | ||
| 10% | 0.9169/0.8908/0.9036 | 0.9087/0.8230/0.8537 | 0.9308/0.9179/0.9243 |
| 20% | 0.9242/0.9002/0.9120 | 0.9089/0.8455/0.8690 | 0.9387/0.9255/0.9321 |
| 50% | 0.9250/0.9137/0.9193 | 0.9114/0.8642/0.8826 | 0.9419/0.9354/0.9386 |
| 100% | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Data size . | Precision/Recall/F1-score . | ||
| 10% | 0.9169/0.8908/0.9036 | 0.9087/0.8230/0.8537 | 0.9308/0.9179/0.9243 |
| 20% | 0.9242/0.9002/0.9120 | 0.9089/0.8455/0.8690 | 0.9387/0.9255/0.9321 |
| 50% | 0.9250/0.9137/0.9193 | 0.9114/0.8642/0.8826 | 0.9419/0.9354/0.9386 |
| 100% | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| ML-Net | 0.8756/0.8142/0.8437 | 0.8364/0.7309/0.7655 | 0.8849/0.8514/0.8678 |
| MPT | 0.8967/0.8624/0.8778 | 0.8670/0.8012/0.8191 | 0.8985/0.8887/0.8931 |
| BioBERT | 0.9343/0.9010/0.9174 | 0.9214/0.8417/0.8725 | 0.9440/0.9254/0.9346 |
| PubMedBERT | 0.9243/0.8946/0.9092 | 0.8933/0.8681/0.8740 | 0.9363/0.9214/0.9288 |
| BioM-S | 0.9214/0.8985/0.9098 | 0.9123/0.8590/0.8822 | 0.9359/0.9240/0.9299 |
| BioM-D | 0.9288/0.8838/0.9058 | 0.8975/0.8461/0.8648 | 0.9427/0.9140/0.9281 |
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| Our method | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| ML-Net | 0.8756/0.8142/0.8437 | 0.8364/0.7309/0.7655 | 0.8849/0.8514/0.8678 |
| MPT | 0.8967/0.8624/0.8778 | 0.8670/0.8012/0.8191 | 0.8985/0.8887/0.8931 |
| BioBERT | 0.9343/0.9010/0.9174 | 0.9214/0.8417/0.8725 | 0.9440/0.9254/0.9346 |
| PubMedBERT | 0.9243/0.8946/0.9092 | 0.8933/0.8681/0.8740 | 0.9363/0.9214/0.9288 |
| BioM-S | 0.9214/0.8985/0.9098 | 0.9123/0.8590/0.8822 | 0.9359/0.9240/0.9299 |
| BioM-D | 0.9288/0.8838/0.9058 | 0.8975/0.8461/0.8648 | 0.9427/0.9140/0.9281 |
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| Our method | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| ML-Net | 0.8756/0.8142/0.8437 | 0.8364/0.7309/0.7655 | 0.8849/0.8514/0.8678 |
| MPT | 0.8967/0.8624/0.8778 | 0.8670/0.8012/0.8191 | 0.8985/0.8887/0.8931 |
| BioBERT | 0.9343/0.9010/0.9174 | 0.9214/0.8417/0.8725 | 0.9440/0.9254/0.9346 |
| PubMedBERT | 0.9243/0.8946/0.9092 | 0.8933/0.8681/0.8740 | 0.9363/0.9214/0.9288 |
| BioM-S | 0.9214/0.8985/0.9098 | 0.9123/0.8590/0.8822 | 0.9359/0.9240/0.9299 |
| BioM-D | 0.9288/0.8838/0.9058 | 0.8975/0.8461/0.8648 | 0.9427/0.9140/0.9281 |
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| Our method | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
| . | Label-based micro-avg. . | Label-based macro-avg. . | Instance-based . |
|---|---|---|---|
| Systems . | Precision/Recall/F1-score . | ||
| ML-Net | 0.8756/0.8142/0.8437 | 0.8364/0.7309/0.7655 | 0.8849/0.8514/0.8678 |
| MPT | 0.8967/0.8624/0.8778 | 0.8670/0.8012/0.8191 | 0.8985/0.8887/0.8931 |
| BioBERT | 0.9343/0.9010/0.9174 | 0.9214/0.8417/0.8725 | 0.9440/0.9254/0.9346 |
| PubMedBERT | 0.9243/0.8946/0.9092 | 0.8933/0.8681/0.8740 | 0.9363/0.9214/0.9288 |
| BioM-S | 0.9214/0.8985/0.9098 | 0.9123/0.8590/0.8822 | 0.9359/0.9240/0.9299 |
| BioM-D | 0.9288/0.8838/0.9058 | 0.8975/0.8461/0.8648 | 0.9427/0.9140/0.9281 |
| Bioformer | 0.9367/0.9002/0.9181 | 0.9038/0.8823/0.8875 | 0.9414/0.9256/0.9334 |
| Our method | 0.9342/0.9062/0.9200 | 0.9155/0.8695/0.8881 | 0.9475/0.9311/0.9392 |
Table 9 presents the classification errors of each topic type with the false positive rate (FPR) and false negative rate (FNR) of the proposed method. It is observed that a relatively high proportion of FPR occurred in ‘Treatment’. This is because more than 40% of data is related to ‘Treatment’, which causes the model to be biased towards the majority class. The imbalanced data issues also affect small classes, such as ‘Epidemic Forecasting’ and ‘Transmission’. Based on our further analysis, we observed that all positive instances of ‘Epidemic Forecasting’ only co-occur with the negative instances of ‘Treatment’. However, the co-occurrence of ‘Transmission’ and ‘Treatment’ is mixed, which causes the proposed model to be more affected by the imbalanced data problem, and therefore, a great portion of FNR occurred in ‘Transmission’. Our error analysis shows that the performance improvement in multi-label classification remains limited, although we have adopted the BCEWithLogitsLoss as the loss function to alleviate the problem of data imbalance. An effective loss function to decrease the impact of imbalanced data issues shall be the foremost issue to be addressed in our future work.
| LABEL (support) . | #FP . | #FN . | FPR . | FNR . |
|---|---|---|---|---|
| Treatment (1035) | 57 | 100 | 6.82% | 5.50% |
| Diagnosis (722) | 41 | 94 | 2.30% | 13.01% |
| Prevention (926) | 45 | 63 | 2.85% | 6.80% |
| Machanism (567) | 20 | 61 | 1.03% | 9.82% |
| Transmission (128) | 9 | 48 | 0.37% | 37.50% |
| Epidemic forecasting (41) | 10 | 5 | 0.40% | 12.19% |
| Case report (197) | 6 | 11 | 0.26% | 5.58% |
| LABEL (support) . | #FP . | #FN . | FPR . | FNR . |
|---|---|---|---|---|
| Treatment (1035) | 57 | 100 | 6.82% | 5.50% |
| Diagnosis (722) | 41 | 94 | 2.30% | 13.01% |
| Prevention (926) | 45 | 63 | 2.85% | 6.80% |
| Machanism (567) | 20 | 61 | 1.03% | 9.82% |
| Transmission (128) | 9 | 48 | 0.37% | 37.50% |
| Epidemic forecasting (41) | 10 | 5 | 0.40% | 12.19% |
| Case report (197) | 6 | 11 | 0.26% | 5.58% |
| LABEL (support) . | #FP . | #FN . | FPR . | FNR . |
|---|---|---|---|---|
| Treatment (1035) | 57 | 100 | 6.82% | 5.50% |
| Diagnosis (722) | 41 | 94 | 2.30% | 13.01% |
| Prevention (926) | 45 | 63 | 2.85% | 6.80% |
| Machanism (567) | 20 | 61 | 1.03% | 9.82% |
| Transmission (128) | 9 | 48 | 0.37% | 37.50% |
| Epidemic forecasting (41) | 10 | 5 | 0.40% | 12.19% |
| Case report (197) | 6 | 11 | 0.26% | 5.58% |
| LABEL (support) . | #FP . | #FN . | FPR . | FNR . |
|---|---|---|---|---|
| Treatment (1035) | 57 | 100 | 6.82% | 5.50% |
| Diagnosis (722) | 41 | 94 | 2.30% | 13.01% |
| Prevention (926) | 45 | 63 | 2.85% | 6.80% |
| Machanism (567) | 20 | 61 | 1.03% | 9.82% |
| Transmission (128) | 9 | 48 | 0.37% | 37.50% |
| Epidemic forecasting (41) | 10 | 5 | 0.40% | 12.19% |
| Case report (197) | 6 | 11 | 0.26% | 5.58% |
The COVID-19 pandemic has had a wide-ranging influence on society, causing increased death and morbidity, as well as interruptions in daily life and overall unease. Many of these issues are unique in terms of type, scope or cause, and one of the most effective methods to solve them is to have better information, that is, the right amount of precise data at the point where it can be implemented (42). However, the difficulty in locating credible and practical knowledge unique to a given context triggered a second epidemic: information overload, which was compounded by the disease’s evolving understanding and a wave of article retractions from even the most prestigious publications. Meanwhile, members of the public were subjected to severe psychological stress as a result of shifting public health policies, severe economic consequences and health uncertainties, all while dealing with their own information overload via news and social media, which was exacerbated by inconsistent messaging and deliberate misinformation campaigns. However, many existing NLP tasks can directly address information requirements during the COVID-19 epidemic, and our proposed method showed the promising results just by improving on existing NLP tasks.
In addition, the establishment of COVID Moonshot and collaboration between COVID Moonshot and PostEra, a startup focusing on medicinal chemistry powered by machine learning, to deliver an antiviral drug for COVID, showcased the potential for drug discovery to be accelerated with the assistance from machine learning. This is beneficial to the world as more breakthroughs may be achieved for more diseases in a shorter duration, bringing possible cures to more people.
Concluding remarks
BioNLP is gaining importance due to the huge yearly increases in the publication of biomedical literature that makes manual curation very challenging. In this research, we introduced a BERT-based ensemble learning approach for the NLM-CHEM and LitCovid tracks in the BioCreative VII Challenge. We explored various state-of-the-art biomedical-specific pre-trained BERT models in both tracks. As the different BERT models have their own characteristics, they also had their distinct advantages which enabled them to perform well. Therefore, by combining them through ensemble learning, the system’s performance can be improved. For the NLM-CHEM track, our model achieved remarkable performance in chemical identification. We further proposed a MeSH ID normalization algorithm for the normalization of chemical entities. The experiment results demonstrated that the dynamic programming-based method is effective in normalizing chemical entities. As for the LitCovid track, our BERT-based ensemble approach achieved state-of-the-art performance in detecting topics in the COVID-19 literature. In addition, this study also explores the performance of various BERT-based models in the NLM-CHEM and LitCovid tasks. We have proved that the integration of BERT models using ensemble learning can further improve the system performance. The results are able to contribute to future research while addressing both tasks.
In the future, deeper semantic information will be integrated into the BERT architecture by exploring other aspects, such as the dependency construction in texts. We will also use relation extraction algorithms to recognize chemical relation passages and construct the relation network of chemicals.
Acknowledgements
This research was supported by the Ministry of Science and Technology of Taiwan under grants MOST 110-2634-F-038-006, MOST 110-2634-F-A49-004, and MOST 109-2410-H-038-012-MY2.
Conflict of interest
None declared.
References
Appendix A
Author notes
First Authors.



