-
PDF
- Split View
-
Views
-
Cite
Cite
Yiding Geng, Lu Jin, Guangjue Tang, Zhangxiang Zhao, Yunyan Gu, Dan Yang, LiqBioer: a manually curated database of cancer biomarkers in body fluid, Database, Volume 2022, 2022, baac077, https://doi.org/10.1093/database/baac077
Close - Share Icon Share
Abstract
Cancer biomarkers are measurable indicators that play vital roles in clinical applications. Biomarkers in body fluids have gained considerable attention since the development of liquid biopsy, and their data volume is rapidly increasing. Nevertheless, current research lacks the compilation of published cancer body fluid biomarkers into a centralized and sustainable repository for researchers and clinicians, despite a handful of small-scale and specific data resources. To fulfill this purpose, we developed liquid biomarker (LiqBioer) containing 6231 manually curated records from 3447 studies, covering 3056 biomarkers and 74 types of cancer in 22 tissues. LiqBioer allows users to browse and download comprehensive information on body liquid biomarkers, including cancer types, source studies and clinical usage. As a comprehensive resource for body fluid biomarkers of cancer, LiqBioer is a powerful tool for researchers and clinicians to query and retrieve biomarkers in liquid biopsy.
Introduction
Biomarkers are biochemical, cellular, gene-related or molecular indicators of diseases that are measurable in physical media, such as blood, body fluids, tissues or cells (1). Biomarkers are widely used in clinical applications including early cancer diagnosis, improved cancer prognosis, early detection of cancer relapse, real-time monitoring of therapeutic efficacy and therapeutic resistance (2).
Cancer is a complex disease with a high mortality rate and is a major global public health concern (3, 4). Biomarkers have been widely used for cancer diagnosis and treatment since their development (5). In the era of precision oncology, molecular profiles of individual cancer patients can facilitate treatment decisions, monitor subsequent responses and alert patients to the emergence of therapeutic resistance and relapse. Gold standard molecular profiling is performed for resected or biopsied tumors (6). However, tumor biopsy is invasive, inconvenient and difficult to track in terms of temporal and spatial heterogeneity (7).
In the last two decades, advances in liquid biopsy have paved the way for tumor tracking (8). The management and prognosis of patients will also benefit from the development of liquid biopsy technology (9, 10). Compared with tissue-based biopsy approaches, liquid biopsy is less invasive and samples are more easily obtainable during disease progression, thus complementing the development of dynamic molecular imaging assessments (11). Liquid biopsy tests include circulating tumor cells (CTCs), circulating nucleic acids, including cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) in the plasma, cell-free RNAs (mRNAs, long non-coding RNAs and microRNAs), extracellular vesicles, tumor-educated platelets, proteins and metabolites that can be found in a range of body fluids (12, 13).
Clinically, the Food and Drug Administration approved CTC enumeration using CellSearch technology to stratify patients with breast, colorectal and prostate cancers (14). ctDNA analysis has been optimized for routine diagnosis in disease monitoring and the identification of actionable mutations (15, 16). For example, the detection of EGFR mutations in ctDNA in non-small cell lung cancer indicates that patients are ready for targeted therapy (17). Recent studies have shown that exosomes are effective in liquid biopsy for early diagnosis, disease monitoring and prognosis prediction (18–20). CTC, cfDNA and extracellular vesicles are currently the most commonly used tests in clinical practice.
Although many online tumor biomarker databases are available, they are used for tumor biopsies. For example, Liver Cancer Expression Resource (21) and Colorectal Cancer Biomarker Database (22). There are a few liquid biopsy databases, such as the Protein Body Fluid Database: human body fluid proteome (HBFP) (23). Nevertheless, HBFP contains only information about proteins and is not specific to cancer. Thus, we developed a database of liquid biomarker (LiqBioer) that covers multiple types of biomarkers for different types of cancers.
Result
Data collection
We extracted body fluid biomarkers of cancer from the literature. We searched the published literature before 28 February 2022, from the PubMed database with keywords such as ‘body fluid’, ‘liquid biopsy’, ‘circulating tumor cells’, ‘circulating DNA’, ‘circulating RNA’, ‘carcinoembryonic antigen’, ‘extracellular vesicle’, ‘exosome’, ‘ectosome’, ‘shedding microvesicle’, ‘apoptotic body’ and ‘secretome’, removing reviews, meta-analysis papers and papers without full text. We used R programming language to filter out abstracts that did not contain the ‘cancer’ word. Subsequently, we manually searched the abstracts for literature containing biomarker names, cancer types and body fluid sources. We collected drug target, sensitivity and resistance data of biomarkers from the collected papers. Figure 1A shows the common types of body fluids and biomarkers. The first two parts of Figure 1B schematically illustrate the data-collection process.
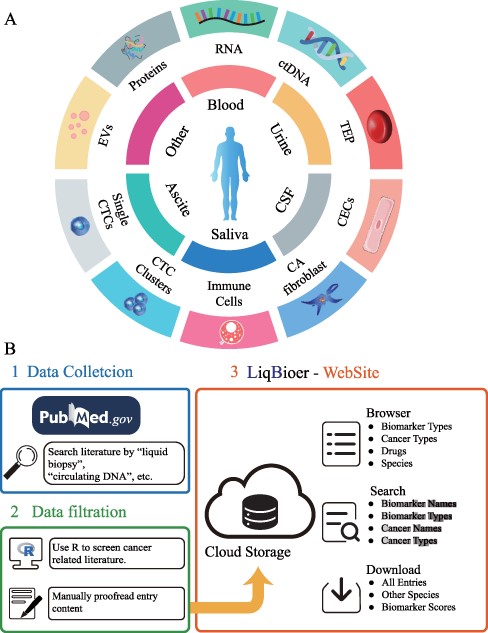
Construction of LiqBioer. (A) Biomarkers in body fluids, such as blood, urine, CSF, saliva and ascites. EVs, proteins, circulating cell-free RNA (non-coding and messenger RNA), ctDNA, TEPs, the tumor cell fraction with CTCs and the non-tumor cell fraction (immune cells, CECs or CA fibroblasts) can be detected in the different body fluids. CSF, cerebrospinal fluid; EV, extracellular vesicle; CA fibroblast, cancer-associated fibroblast; CEC, circulating endothelial cell; TEP, tumor-educated platelet. (B) Process of data collection, filtering and website structure. 1: Data collection. 2: Data filtration. 3: Structure of website.
Database and web interface design
LiqBioer is freely available without registration. LiqBioer was designed to operate optimally with Google Chrome and Firefox. The web interface was implemented using Java technology (Java Server Pages, Asynchronous JavaScript, and Ajax), Google Code API and XML under the integrated development environment Eclipse (https://www.eclipse.org/). Public access to the database was strictly read-only. The web application was hosted and deployed on the Apache Tomcat server (version 9). LiqBioer provides a user-friendly web interface that allows for intuitive browsing and searching. The website includes five primary functional pages: ‘Home’, ‘Browser’, ‘Search’, ‘Download’ and ‘Help’. The third part of Figure 1B shows the structure of LiqBioer. Figure 2 illustrates the pages and how to use the browse and search functions.
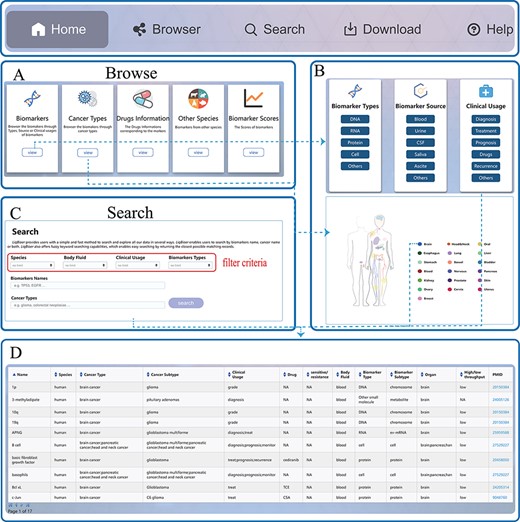
Usage instructions of LiqBioer. (A) Browse page. (B) Subpages of the browse page. (C) Search page. (D) Table of search and browse results.
The ‘Home’ page provides a speedy search and synopsis of the database LiqBioer. Users can input the name of a biomarker or the type of cancer to retrieve the corresponding information.
The ‘Browse’ page provides a browse through modules of all records. The ‘Browse’ page contains five modules, ‘biomarker’, ‘cancer types’, ‘other species’, ‘drugs’ and ‘scores’. The ‘biomarker’ module includes the types, sources and clinical usage of biomarkers. The ‘cancer’-type module displays the primary sites of cancer. Users can click on the buttons of organs or icons on the human body to browse for cancer biomarkers of specific organs.
The ‘Search’ page allows users to set four search conditions: species, source of body fluid, clinical usage and types of biomarkers. Subsequently, the users can search for biomarkers using the name of a biomarker and the type of cancer.
The ‘Download’ page provides the latest data on body fluid biomarkers of cancer. Users can download four files containing all the information for all markers. Other-Species.txt contains information on the biomarkers of species other than humans. Drugs.txt contains information about biomarkers that contain information about drugs. Scores.txt contains the clinical scores of the biomarkers.
The ‘Help’ page guides users to use the website, learn the website’s statistics and contact us by e-mail. Users can obtain statistics through pie charts designed using HighCharts (a JavaScript framework for drawing statistical charts). Pie charts allow users to click on sectors to see the details of the charts. If users have any questions or suggestions, they can send us an e-mail by clicking on ‘contact us’.
Clinical scores of biomarkers
The LiqBioer database was used to calculate a statistical score to evaluate the clinical confidence of the body fluid biomarkers of cancer. The clinical score was assessed based on the following four aspects: presence/absence of a drug in that entry (|$S1 = \left\{ {1, 0} \right\}$|), presence/absence of information on drug resistance (|$S2 = \left\{ {1, 0} \right\}$|), high-throughput/low-throughput/absence of detection techniques (|$S3 = \left\{ {0.2, 1, 0} \right\}$|) and the number of papers containing the biomarker (|$n$|). In summary, a high clinical score indicates detailed medication information and low-throughput biological experiments. Clinical scores were calculated as follows:
Table 1 shows the 10 biomarkers with the highest clinical scores.
| Biomarkers . | Score . |
|---|---|
| miR-342-3p | |$5.854 \times {10^3}$| |
| C/EBPβ | |$5.854 \times {10^3}$| |
| MUC5AC | |$5.854 \times {10^3}$| |
| circ_0007385 | |$5.853 \times {10^3}$| |
| sTn | |$5.853 \times {10^3}$| |
| PHEX-AS1 | |$5.852 \times {10^3}$| |
| RP11-77G23.5 | |$5.851 \times {10^3}$| |
| GOLPH3 | |$5.850 \times {10^3}$| |
| circTRPS1 | |$5.850 \times {10^3}$| |
| miR141-3p | |$5.850 \times {10^3}$| |
| Biomarkers . | Score . |
|---|---|
| miR-342-3p | |$5.854 \times {10^3}$| |
| C/EBPβ | |$5.854 \times {10^3}$| |
| MUC5AC | |$5.854 \times {10^3}$| |
| circ_0007385 | |$5.853 \times {10^3}$| |
| sTn | |$5.853 \times {10^3}$| |
| PHEX-AS1 | |$5.852 \times {10^3}$| |
| RP11-77G23.5 | |$5.851 \times {10^3}$| |
| GOLPH3 | |$5.850 \times {10^3}$| |
| circTRPS1 | |$5.850 \times {10^3}$| |
| miR141-3p | |$5.850 \times {10^3}$| |
| Biomarkers . | Score . |
|---|---|
| miR-342-3p | |$5.854 \times {10^3}$| |
| C/EBPβ | |$5.854 \times {10^3}$| |
| MUC5AC | |$5.854 \times {10^3}$| |
| circ_0007385 | |$5.853 \times {10^3}$| |
| sTn | |$5.853 \times {10^3}$| |
| PHEX-AS1 | |$5.852 \times {10^3}$| |
| RP11-77G23.5 | |$5.851 \times {10^3}$| |
| GOLPH3 | |$5.850 \times {10^3}$| |
| circTRPS1 | |$5.850 \times {10^3}$| |
| miR141-3p | |$5.850 \times {10^3}$| |
| Biomarkers . | Score . |
|---|---|
| miR-342-3p | |$5.854 \times {10^3}$| |
| C/EBPβ | |$5.854 \times {10^3}$| |
| MUC5AC | |$5.854 \times {10^3}$| |
| circ_0007385 | |$5.853 \times {10^3}$| |
| sTn | |$5.853 \times {10^3}$| |
| PHEX-AS1 | |$5.852 \times {10^3}$| |
| RP11-77G23.5 | |$5.851 \times {10^3}$| |
| GOLPH3 | |$5.850 \times {10^3}$| |
| circTRPS1 | |$5.850 \times {10^3}$| |
| miR141-3p | |$5.850 \times {10^3}$| |
Statistics of LiqBioer
LiqBioer contains 6231 entries of biomarkers among 3056 biomarkers for 74 types of cancer in 22 tissues from 3447 articles. The biomarkers were obtained from 14 body fluids, including blood, ascites, urine and saliva, encompassing DNA, RNA, protein, cell and other types. The LiqBioer database also contains species other than humans, such as mouse, canine, feline and gallus (Figure 3). Figure 3A shows the distribution of biomarker types; more than half of them were RNA biomarkers. Figure 3B shows that the tissue sites and the breast tissue had the most biomarkers in body fluids. Figure 3C shows the distribution of body fluid types and the blood accounted for the vast majority of cases. Figure 3D shows the species distribution; the human species are the majority of all species. Figure 3E shows the distribution of cancer types; breast cancer tops the list, followed by lung cancer.
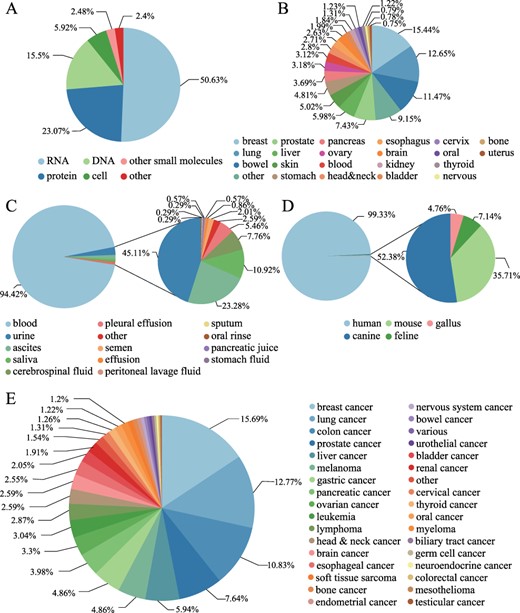
Statistics of biomarkers in LiqBioer. (A) Classification of biomarkers. (B) Tissues of cancer. (C) Source of body fluid. (D) Source species for biomarkers. (E) Cancer types for biomarkers.
Discussion and conclusion
LiqBioer is a manually curated database that contains multiple types of body fluid cancer biomarkers. LiqBioer can assist clinicians in diagnosis and scientific researchers in selecting the research objectives. LiqBioer will be updated regularly according to the newly available biomarkers and user-submitted data as an interactive website.
However, LiqBioer only collected data and did not design the analytics for further investigation. Analysis tools and other functional modules will be available for subsequent updates. A module on clinical guidelines and clinical trials will be added to enable clinicians in accessing information more effectively.
In conclusion, LiqBioer will be a valuable tool for researchers and clinicians to retrieve biomarkers for liquid biopsy of tumors and manage cancer treatment.
Abbreviations
cfDNA: tumor-derived fraction of cell-free DNA
ctDNA: circulating tumor DNA
CA fibroblast: cancer-associated fibroblast
CEC: circulating endothelial cell
CTC: circulating tumor cell
EV: extracellular vesicle
LiqBioer: Liquid Biomarker
TEP: tumor-educated platelet
Conflict of interest
The authors declare that they have no competing interests.
References
Author notes
contributed equally to this work.



