-
PDF
- Split View
-
Views
-
Cite
Cite
Jiaqi Chen, Zhengxin Chen, Rufei Chen, Dehua Feng, Tianyi Li, Huirui Han, Xiaoman Bi, Zhenzhen Wang, Kongning Li, Yongsheng Li, Xia Li, Limei Wang, Jin Li, HCDT: an integrated highly confident drug–target resource, Database, Volume 2022, 2022, baac101, https://doi.org/10.1093/database/baac101
Close - Share Icon Share
Abstract
Drug–target association plays an important role in drug discovery, drug repositioning, drug synergy prediction, etc. Currently, a lot of drug-related databases, such as DrugBank and BindingDB, have emerged. However, these databases are separate, incomplete and non-uniform with different criteria. Here, we integrated eight drug-related databases; collected, filtered and supplemented drugs, target genes and experimentally validated (highly confident) associations and built a highly confident drug–target (HCDT: http://hainmu-biobigdata.com/hcdt) database. HCDT database includes 500 681 HCDT associations between 299 458 drugs and 5618 target genes. Compared to individual databases, HCDT database contains 1.1 to 254.2 times drugs, 1.8–5.5 times target genes and 1.4–27.7 times drug–target associations. It is normative, publicly available and easy for searching, browsing and downloading. Together with multi-omics data, it will be a good resource in analyzing the drug functional mechanism, mining drug-related biological pathways, predicting drug synergy, etc.
Database URL: http://hainmu-biobigdata.com/hcdt
Introduction
Drug discovery is a time-consuming, costly and risky process (1). According to a report published in 2021, the average time to develop a new drug from clinical trials to market was 7.5 years (2). With clinical development proceeding from Phase 1 to Phases 2 and 3, the average cost per study increased for all therapeutic areas. However, the average success rate for developing new drugs was only 2.01% (2, 3). The annual investment in drug development has gradually increased, even while the number of Food and Drug Administration-approved drugs has been declining since 1995 (4). Between 2009 and 2018, the median cost of developing a new drug was $985 million, while the average sum totaled $1.3 billion. In all therapeutic areas, oncology and immunomodulatory drugs were the most expensive to develop, coming in at a median of $2.8 billion and a mean of $4.5 billion (5).The cost of new drug development will continue to grow. Hence, it is urgent to find a new strategy to discover drugs (1).
Drug repositioning (6), also known as drug repurposing, uses the molecular structure, indications and adverse effects of a known drug to develop new functions beyond the drug testing, safety review and clinical phases of the traditional drug development model. It reduces costs and time with low risk (1, 7).
Using the structure, properties and target interactions to predict new functions of drugs is a key step in drug repositioning, and a key point is to know and understand the relationship between drugs and targets (8, 9). The association between a drug and target gene can be determined by in vitro and in vivo studies, and a lot of drug-related databases have been constructed, such as ChEMBL (10), DrugBank (11) and DtoPdb (12). A variety of computational methods to predict drug–target association have been proposed, such as data mining (13), machine learning methods (14, 15), neural networks (13, 16) and collaborative matrix decomposition methods (9). Key information used in these methods is the known experimentally confirmed drug–target associations.
In some drug-related databases, the associations between drugs and targets have been experimentally validated, such as DrugBank, KEGG (17) and PubChem (18). However, in some databases, in addition to experimentally confirmed drug–target association, there are also computational predicted drug–target associations, such as BindingDB (19), STITCH (20) and ChEMBL. We can get quite different drug–target associations when using different predicting methods or thresholds. Therefore, experimentally validated drug–target associations are more confident than the predicted ones and much useful in drug repositioning and other studies.
The information in these databases varies a lot due to their different research purposes. SIDER and DrugBank focus on the drug structure and function, with less information on target genes (11, 21). DGIdb focuses more on drug–target associations without specific information on corresponding drugs and targets (12). STITCH and SuperPred focus on the prediction of new drug functions (20, 22). PharmGKB contains too little information on experimentally validated data, while BindingDB has too much information on predicted data to meet the data needs of researchers (19, 23). Meanwhile, there are different identifications for drugs and genes, and it makes extra burden for researchers when they acquire information from different databases.
Here, we used unified standards for drugs and target genes, integrated the experimentally validated drug–target associations in the current popular drug-related databases and constructed a highly confident drug–target (HCDT (http://hainmu-biobigdata.com/hcdt) database.
Materials and methods
Data collection and processing
We have collected 13 commonly used drug databases: BindingDB, ChEMBL, DGIdb, DrugBank, GtoPdb, PharmGKB, PubChem, TTD (24), CancerDR (25), STITCH, SIDER, SuperPred and KEGG (see details in Table 1). The drug-related data are expected to contain simplified molecular input line entry system (SMILES), International Union of Pure and Applied Chemists (IUPAC) name, International Chemical Identifier (INCH), The Anatomical Therapeutic Chemical codes and binding affinity of the drug; for the gene data, it is guaranteed to contain one of the gene symbols, Entrez ID, Ensembl ID or UniProt ID, which can be mapped with the gene information in the HGNC database.
| . | Drug label . | Gene label . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Database . | SMILES . | IUPAC name . | Inch . | ATC codes . | Binding affinity . | Gene symbol . | Entrez ID . | Ensembl ID . | UniProt ID . | Criteria . |
| BindingDB ChEMBL GtoPdb PubChem TTD | √ √ √ √ √ | √ √ √ | √ √ √ √ √ | √ √ | √ √ √ √ √ | √ √ √ √ | | √ √ √ | √ √ √ √ √ | Including criteria (i) binding affinity, including at least one of Ki, Kd, IC50 or EC50 ≤ 10 μM; (ii) the presence of UniProt ID representation of the protein and (iii) the protein being tagged as ‘review’. |
| DGIdb | √ | √ | √ | √ | Fourteen of these experimentally validated database sources are screened for drug–target interactions and eight are excluded. See the details in Supplementary Table S1. | |||||
| DrugBank | √ | √ | √ | √ | √ | √ | √ | Interactions with no clear target information are excluded. | ||
| PharmGKB | √ | √ | √ | √ | √ | √ | Only the data marked as ‘associated’ are included. | |||
| CancerDR | √ | √ | √ | CancerDR database is excluded as it is mainly used for the study of drugs and sensitive cell lines, but lacks the information of the corresponding target effects. | ||||||
| STITCH SuperPred | √ √ | √ | √ | √ | STITCH and SuperPred are excluded, as the majority of drug–target association are based on computational prediction, but not confirmed by biological experiments, which do not meet the high confident purpose of this study. | |||||
| SIDER | √ | √ | SIDER database is excluded as it is used for the study of drug side effects but lacks target information. | |||||||
| KEGG | √ | √ | √ | √ | √ | √ | √ | KEGG is excluded, as the drug–target information is simply collected from ChEMBL, Drugbank and PubChem, without selection or integration. | ||
| . | Drug label . | Gene label . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Database . | SMILES . | IUPAC name . | Inch . | ATC codes . | Binding affinity . | Gene symbol . | Entrez ID . | Ensembl ID . | UniProt ID . | Criteria . |
| BindingDB ChEMBL GtoPdb PubChem TTD | √ √ √ √ √ | √ √ √ | √ √ √ √ √ | √ √ | √ √ √ √ √ | √ √ √ √ | | √ √ √ | √ √ √ √ √ | Including criteria (i) binding affinity, including at least one of Ki, Kd, IC50 or EC50 ≤ 10 μM; (ii) the presence of UniProt ID representation of the protein and (iii) the protein being tagged as ‘review’. |
| DGIdb | √ | √ | √ | √ | Fourteen of these experimentally validated database sources are screened for drug–target interactions and eight are excluded. See the details in Supplementary Table S1. | |||||
| DrugBank | √ | √ | √ | √ | √ | √ | √ | Interactions with no clear target information are excluded. | ||
| PharmGKB | √ | √ | √ | √ | √ | √ | Only the data marked as ‘associated’ are included. | |||
| CancerDR | √ | √ | √ | CancerDR database is excluded as it is mainly used for the study of drugs and sensitive cell lines, but lacks the information of the corresponding target effects. | ||||||
| STITCH SuperPred | √ √ | √ | √ | √ | STITCH and SuperPred are excluded, as the majority of drug–target association are based on computational prediction, but not confirmed by biological experiments, which do not meet the high confident purpose of this study. | |||||
| SIDER | √ | √ | SIDER database is excluded as it is used for the study of drug side effects but lacks target information. | |||||||
| KEGG | √ | √ | √ | √ | √ | √ | √ | KEGG is excluded, as the drug–target information is simply collected from ChEMBL, Drugbank and PubChem, without selection or integration. | ||
| . | Drug label . | Gene label . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Database . | SMILES . | IUPAC name . | Inch . | ATC codes . | Binding affinity . | Gene symbol . | Entrez ID . | Ensembl ID . | UniProt ID . | Criteria . |
| BindingDB ChEMBL GtoPdb PubChem TTD | √ √ √ √ √ | √ √ √ | √ √ √ √ √ | √ √ | √ √ √ √ √ | √ √ √ √ | | √ √ √ | √ √ √ √ √ | Including criteria (i) binding affinity, including at least one of Ki, Kd, IC50 or EC50 ≤ 10 μM; (ii) the presence of UniProt ID representation of the protein and (iii) the protein being tagged as ‘review’. |
| DGIdb | √ | √ | √ | √ | Fourteen of these experimentally validated database sources are screened for drug–target interactions and eight are excluded. See the details in Supplementary Table S1. | |||||
| DrugBank | √ | √ | √ | √ | √ | √ | √ | Interactions with no clear target information are excluded. | ||
| PharmGKB | √ | √ | √ | √ | √ | √ | Only the data marked as ‘associated’ are included. | |||
| CancerDR | √ | √ | √ | CancerDR database is excluded as it is mainly used for the study of drugs and sensitive cell lines, but lacks the information of the corresponding target effects. | ||||||
| STITCH SuperPred | √ √ | √ | √ | √ | STITCH and SuperPred are excluded, as the majority of drug–target association are based on computational prediction, but not confirmed by biological experiments, which do not meet the high confident purpose of this study. | |||||
| SIDER | √ | √ | SIDER database is excluded as it is used for the study of drug side effects but lacks target information. | |||||||
| KEGG | √ | √ | √ | √ | √ | √ | √ | KEGG is excluded, as the drug–target information is simply collected from ChEMBL, Drugbank and PubChem, without selection or integration. | ||
| . | Drug label . | Gene label . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Database . | SMILES . | IUPAC name . | Inch . | ATC codes . | Binding affinity . | Gene symbol . | Entrez ID . | Ensembl ID . | UniProt ID . | Criteria . |
| BindingDB ChEMBL GtoPdb PubChem TTD | √ √ √ √ √ | √ √ √ | √ √ √ √ √ | √ √ | √ √ √ √ √ | √ √ √ √ | | √ √ √ | √ √ √ √ √ | Including criteria (i) binding affinity, including at least one of Ki, Kd, IC50 or EC50 ≤ 10 μM; (ii) the presence of UniProt ID representation of the protein and (iii) the protein being tagged as ‘review’. |
| DGIdb | √ | √ | √ | √ | Fourteen of these experimentally validated database sources are screened for drug–target interactions and eight are excluded. See the details in Supplementary Table S1. | |||||
| DrugBank | √ | √ | √ | √ | √ | √ | √ | Interactions with no clear target information are excluded. | ||
| PharmGKB | √ | √ | √ | √ | √ | √ | Only the data marked as ‘associated’ are included. | |||
| CancerDR | √ | √ | √ | CancerDR database is excluded as it is mainly used for the study of drugs and sensitive cell lines, but lacks the information of the corresponding target effects. | ||||||
| STITCH SuperPred | √ √ | √ | √ | √ | STITCH and SuperPred are excluded, as the majority of drug–target association are based on computational prediction, but not confirmed by biological experiments, which do not meet the high confident purpose of this study. | |||||
| SIDER | √ | √ | SIDER database is excluded as it is used for the study of drug side effects but lacks target information. | |||||||
| KEGG | √ | √ | √ | √ | √ | √ | √ | KEGG is excluded, as the drug–target information is simply collected from ChEMBL, Drugbank and PubChem, without selection or integration. | ||
Five of these databases were excluded in the following steps: CancerDR database is mainly used for the study of drugs and sensitive cell lines but lacks the information of the corresponding target effects; in STITCH and SuperPred, the majority of drug–target associations are based on computational prediction, but not confirmed by biological experiments, which do not meet the highly confident purpose of this study; SIDER database is used for the study of drug side effects but lacks target information; In KEGG, the drug–target information is simply collected from ChEMBL, DrugBank and PubChem, without selection or collation.
The left eight databases were selected as the original data sources for the HCDT database. To ensure the high confidence of drug–target associations, we used the following criteria: for databases with drug binding affinity information (BindingDB, ChEMBL, GtoPdb, PubChem and TTD), we retained only drug–target associations that met the following three criteria (22): (i) binding affinity, including at least one of Ki, Kd, half maximal inhibitory concentration (IC50) or half maximal effective concentration (EC50) ≤ 10 μM; (ii) the presence of UniProt ID representation of the protein and (iii) the protein being tagged as ‘review’. DGIdb is integrated by several databases. In the latest release of DGIdb 4.0 (26), we screened 14 databases for experimentally validated drug–target association and excluded three databases with drug–target prediction and five databases (ChEMBL, DrugBank, GtoPdb, PharmGKB and TTD) that had been collected separately (see the details in Supplementary Table S1). In PharmGKB, there were three types of drug–target relationships: ‘associated’, ‘not associated’ and ‘ambiguous’, and only the data marked as ‘associated’ were selected. All drug–target associations are validated by in vivo experiments and guaranteed to be of human origin but not of other species.
Drug classifications
There are various kinds of drug classifications in these databases. A drug may be marked as different types in different databases. For example, tisagenlecleucel (27), an immune agent for B-cell lymphoma, is marked as ‘antibody’ in ChEMBL and ‘biotech’ in BindingDB. There are about 268k drugs in BindingDB, which account for 89.5% of drugs in HCDT database. Therefore, we classified drugs based on BindingDB first and then referred other classifications in other databases. There are six drug types in BindingDB: small-molecule organic compounds (synthetic small molecule), synthetic products of biotechnology (Biotech), metabolites, oligomer and combination of drug pairs and unknown drug types that do not fall into the above five categories. For Biotech, there are subtypes such as protein, cell, enzyme, gene in ChEMBL, antibody in ChEMBL and GtoPdb. For oligomer, there are four subtypes, such as oligonucleotide, oligopeptide and oligosaccharide in ChEMBL and peptide in GtoPdb. There are also two special drug types in GtoPdb. For example, apigenina (28), a flavonoid, is shown as a natural product in ChEMBL and GtoPdb, but does not belong to any six types in BindingDB; nitric oxide, a selective pulmonary vasodilator used to treat various forms of pulmonary hypertension in order to improve oxygenation levels, is marked as inorganic in GtoPdb, but it does not belong to any six types in BindingDB. Finally, we got eight types of drugs (Figure 1).
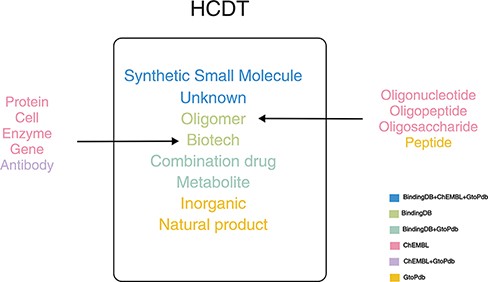
For each drug, we use the label in BindingDB if a drug exists in BindingDB (for example, tisagenlecleucel is marked as ‘biotech’ as it is marked as ‘biotech’ in BindingDB); we match label in the original database to the corresponding BindingDB label if a drug does not exist in BindingDB, but the labels can match [for example, MIPOMERSEN (29) is marked as ‘Oligomer’ as it is marked as ‘Oligonucleotide’ in ChEMBL]; we use label inorganic or natural product, if a drug belonged to these two types.
Gene types
Based on the HGNC database, genes are classified into four groups according to function (30): genes that encode proteins, genes that do not encode ribonucleic acid (RNA), pseudogenes that have no actual function and the remaining genes whose function is not yet clear. Then, they are further subdivided into a total of 24 subtypes based on their specific attached functional objects. The main type of protein-coding gene group is a collection of genes that can encode proteins; the non-coding RNA group is divided into 10 types according to the RNA type that can be transcribed and translated into sex chromosomes, long-stranded non-coding RNA, microRNA and small RNA; pseudogenes are non-functional residues formed during gene evolution and can be divided into three types: T-cell receptor pseudogenes, immunoglobulin pseudogenes and pseudogenes with unknown receptors; for the other 10 subtypes such as functional T-cell receptor genes, complex site component genes and endogenous retroviral genes, they make up the fourth group because the attachment object loci are still unclear.
Data integration
The SMILES information is used as the unique identifier (31) for drugs and then other features, such as IUPAC name (32), and synonyms are matched to the drugs. For genes, the gene symbol is used as the unique identifier and the HGNC database is used to supplement the information on UniProt ID, Entrez ID, gene type, location and Ensembl ID. The drug–target relationships in each database were then integrated based on SMILES and gene symbol.
Results
Data in HCDT database
In total, 299 458 drugs, 5618 genes and 500 681 pairs of associations were obtained in HCDT database (Table 2). BindingDB is the largest source of HCDT database.
| Database . | Number of drugs . | Number of targets . | Number of associations . |
|---|---|---|---|
| BindingDB | 268 001 | 2293 | 357 695 |
| ChEMBL | 17 110 | 1018 | 42 571 |
| DGIdb | 10 025 | 2344 | 41 924 |
| DrugBank | 6393 | 3168 | 27 111 |
| Gtopdb | 8821 | 1766 | 18 352 |
| PharmGKB | 1178 | 1868 | 18 084 |
| PubChem | 11 049 | 3840 | 64 483 |
| TTD | 27 761 | 2510 | 56 048 |
| HCDT database | 299 458 | 5617 | 500 681 |
| Database . | Number of drugs . | Number of targets . | Number of associations . |
|---|---|---|---|
| BindingDB | 268 001 | 2293 | 357 695 |
| ChEMBL | 17 110 | 1018 | 42 571 |
| DGIdb | 10 025 | 2344 | 41 924 |
| DrugBank | 6393 | 3168 | 27 111 |
| Gtopdb | 8821 | 1766 | 18 352 |
| PharmGKB | 1178 | 1868 | 18 084 |
| PubChem | 11 049 | 3840 | 64 483 |
| TTD | 27 761 | 2510 | 56 048 |
| HCDT database | 299 458 | 5617 | 500 681 |
| Database . | Number of drugs . | Number of targets . | Number of associations . |
|---|---|---|---|
| BindingDB | 268 001 | 2293 | 357 695 |
| ChEMBL | 17 110 | 1018 | 42 571 |
| DGIdb | 10 025 | 2344 | 41 924 |
| DrugBank | 6393 | 3168 | 27 111 |
| Gtopdb | 8821 | 1766 | 18 352 |
| PharmGKB | 1178 | 1868 | 18 084 |
| PubChem | 11 049 | 3840 | 64 483 |
| TTD | 27 761 | 2510 | 56 048 |
| HCDT database | 299 458 | 5617 | 500 681 |
| Database . | Number of drugs . | Number of targets . | Number of associations . |
|---|---|---|---|
| BindingDB | 268 001 | 2293 | 357 695 |
| ChEMBL | 17 110 | 1018 | 42 571 |
| DGIdb | 10 025 | 2344 | 41 924 |
| DrugBank | 6393 | 3168 | 27 111 |
| Gtopdb | 8821 | 1766 | 18 352 |
| PharmGKB | 1178 | 1868 | 18 084 |
| PubChem | 11 049 | 3840 | 64 483 |
| TTD | 27 761 | 2510 | 56 048 |
| HCDT database | 299 458 | 5617 | 500 681 |
Drug–target network
A drug–target network is constructed based on the drug–target association. The distribution of degrees for drug (the number of corresponding targets for a drug) is shown in Figure 2. A drug is associated an average 1.7 experimentally validated target genes, and 220 198 (73.53%) drugs are associated with only the target. Staurosporine (33), an adenosine triphosphate–competitive, non-selective protein kinase inhibitor, has up to 333 targets. Similarly, the distribution of degrees for target (the number of corresponding drugs for a target gene) is shown in Figure 3. A target gene is associated an average 89.1 drugs, and 1214 (21.61%) genes are associated with only one drug. HCRTR2 has 6435 related drugs, which is the largest number (34). The protein encoded by HCRTR2 is a G-protein coupled receptor (GPCR), and it is involved in encoding hypothalamic secretagogues that lend themselves to the regulation of appetite and sleep behavior. GPCRs are the largest family of membrane receptors that are targeted by approved drugs, and approximately 35% of approved drugs target GPCRs (35). The drugs associated with HCRTR2 can be divided into activators and inhibitors. The amide carbon group for the junction is present in the activator. The amide carbonyl of the linker forms a hydrogen bond with H350, whose sidechain also contacts the terminal 1,2,3-triazole moiety and the distal phenyl ring of the core, thereby stabilizing the kinked conformation of HCRTR2. As for inhibitors, all antagonists occupy the bottom-most region of the central cavity, overlapping with the last three residues of HCRTR2 and the portion of a compound containing the sulfonamide and the amide-linked phenyltriazole (36). A list of hub drugs and targets is shown in Supplementary Table S2.
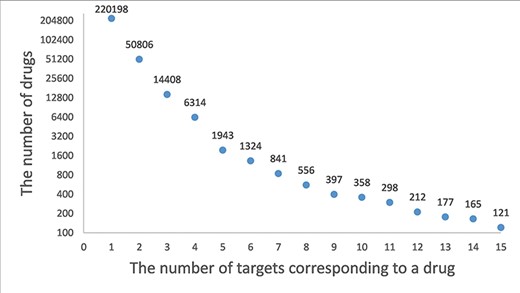
A frequency plot of the number of targets for a drug. The x-axis indicates the number of targets corresponding to a drug and the y-axis indicates the number of drugs. In this figure, only the drugs with less than 15 target genes are shown which account for 99.5% of the drugs.
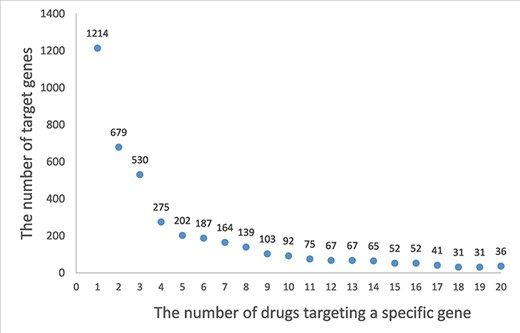
A frequency plot of the number of drugs targeting a specific gene. The x-axis indicates the number of drugs targeting a specific gene, and y-axis indicates the number of target genes. In this figure, only the genes with less than 20 related drugs are shown which account for 73% of the target genes.
We take the top 5% of drugs and genes of degrees in the networks as hubs, where the drugs had at least four targets and the genes had at least 31 target drugs. Then, we compare the hubs between HCDT database and the eight individual databases. For genes, all hub genes in BindingDB and ChEMBL belong to HCDT database (Supplementary Figure S1A and B), which is because BindingDB and ChEMBL provide most drugs for HCDT database. Most hub genes (204/282) in HCDT database are hubs in these individual databases (Supplementary Figure S1I). For drugs, all or most hub drugs in seven individual databases except BindingDB are hubs in HCDT database (Supplementary Figure S2B-H), and about half (7305/13 400) of hub drugs in BindingDB are hubs in HCDT database (Supplementary Figure S2A). These results show that HCDT database is consistent with these individual databases and provides more information as an integrated database.
Drug and target classification
299 458 drugs were classified into eight types in HCDT database (Table 3). There are 268 722 synthetic small-molecule drugs, which is in line with the reality of drug design, where small organic compounds are mostly used in clinical drug development (37). There are only 156 inorganic drugs. Inorganic drugs have been identified as significant candidates for new cancer therapeutic modalities because of their biocompatibility, easy functionalization and fabrication, optical tunable characteristics and chemical stability. However, the problems of eliminating long-term toxicity from metals in vivo and transport of drug carriers have led to the still low number of inorganic drugs entering clinical usage (38, 39).
| Drug type . | Number of drugs . |
|---|---|
| Synthetic small molecule | 268 722 |
| Natural product–derived | 18 864 |
| Biotech | 4978 |
| Oligomer | 2824 |
| Metabolite | 1778 |
| Inorganic | 156 |
| Combination drug | 439 |
| Unknown | 1697 |
| Total | 299 458 |
| Drug type . | Number of drugs . |
|---|---|
| Synthetic small molecule | 268 722 |
| Natural product–derived | 18 864 |
| Biotech | 4978 |
| Oligomer | 2824 |
| Metabolite | 1778 |
| Inorganic | 156 |
| Combination drug | 439 |
| Unknown | 1697 |
| Total | 299 458 |
| Drug type . | Number of drugs . |
|---|---|
| Synthetic small molecule | 268 722 |
| Natural product–derived | 18 864 |
| Biotech | 4978 |
| Oligomer | 2824 |
| Metabolite | 1778 |
| Inorganic | 156 |
| Combination drug | 439 |
| Unknown | 1697 |
| Total | 299 458 |
| Drug type . | Number of drugs . |
|---|---|
| Synthetic small molecule | 268 722 |
| Natural product–derived | 18 864 |
| Biotech | 4978 |
| Oligomer | 2824 |
| Metabolite | 1778 |
| Inorganic | 156 |
| Combination drug | 439 |
| Unknown | 1697 |
| Total | 299 458 |
5618 target genes are classified into 11 subtypes in HCDT database (Table 4), and 5492 of them (97.8%) are protein-coding genes. Different types of genes have different functional mechanisms in drug effect, and HCDT database could be used to analyze the potential mechanism for drugs.
| Gene type . | Number of genes . |
|---|---|
| Gene with protein product | 5492 |
| Pseudogene | 39 |
| RNA, micro | 28 |
| Immunoglobulin gene | 21 |
| RNA, long non-coding | 18 |
| Complex locus constituent | 9 |
| T cell receptor gene | 3 |
| Readthrough | 4 |
| RNA, small nucleolar | 2 |
| RNA, misc | 1 |
| RNA, ribosomal | 1 |
| Total | 5618 |
| Gene type . | Number of genes . |
|---|---|
| Gene with protein product | 5492 |
| Pseudogene | 39 |
| RNA, micro | 28 |
| Immunoglobulin gene | 21 |
| RNA, long non-coding | 18 |
| Complex locus constituent | 9 |
| T cell receptor gene | 3 |
| Readthrough | 4 |
| RNA, small nucleolar | 2 |
| RNA, misc | 1 |
| RNA, ribosomal | 1 |
| Total | 5618 |
| Gene type . | Number of genes . |
|---|---|
| Gene with protein product | 5492 |
| Pseudogene | 39 |
| RNA, micro | 28 |
| Immunoglobulin gene | 21 |
| RNA, long non-coding | 18 |
| Complex locus constituent | 9 |
| T cell receptor gene | 3 |
| Readthrough | 4 |
| RNA, small nucleolar | 2 |
| RNA, misc | 1 |
| RNA, ribosomal | 1 |
| Total | 5618 |
| Gene type . | Number of genes . |
|---|---|
| Gene with protein product | 5492 |
| Pseudogene | 39 |
| RNA, micro | 28 |
| Immunoglobulin gene | 21 |
| RNA, long non-coding | 18 |
| Complex locus constituent | 9 |
| T cell receptor gene | 3 |
| Readthrough | 4 |
| RNA, small nucleolar | 2 |
| RNA, misc | 1 |
| RNA, ribosomal | 1 |
| Total | 5618 |
Comparison with individual databases
HCDT database is an integrated normative database, and it includes as much information as possible for drugs and target genes. For each drug, there are SMILES, PubChem Compound ID, IUPAC name, synonyms, INCH and drug type. For each gene, there are gene symbol, Ensembl ID, gene type, location, Entrez ID and UniPort ID. Not every information exists in these individual databases. For example, in ChEMBL, the target genes are represented by gene-encoded proteins, but not gene symbols; similarly, SMILES and IUPAC name information of some drugs in BindingDB are missing. Therefore, HCDT database is much convenient for researchers.
Compared to individual databases, HCDT database has a much greater volume. It contains 1.1–254.2 times drugs, 1.8–5.5 times target genes and 1.4–27.7 times drug–target associations. Among these databases, BindingDB is the largest data provider, which accounts for 89.50% of drugs, 40.82% of target genes and 71.44% of drug–target associations. For individual databases, there are different types of data information for drugs, targets and associations depending on the purpose of the study. For example, DrugBank lacks target structure information; DGIPdb has descriptions of associations but lacks drug-specific information; BindingDB has records of both drug and target information but is more complicated to operate and has a lot of prediction information. HCDT database combines the advantages of each database and complements the drug, target and association information.
In HCDT database, 399 429 (79.78%) drug–target associations came from only one raw database, most associations (99.21%) existed in less than four raw databases and only 48 associations were present in all eight databases (Figure 4). For example, amitriptyline is a commonly used antidepressant in various types of depression as well as in chronic pain (40). The association of amitriptyline with CACNA1C is only recorded in BindingDB, the association of amitriptyline with Potassium Voltage-Gated Channel Subfamily H Member 2 is recorded in BindingDB, DrugBank and PubChem and the association of amitriptyline with SLC6A4 is recorded in all eight databases. This indicates that the drug–target associations in each database are somewhat related but have some variability due to the different purposes of each database. Integration of these databases in HCDT database greatly improves the volume and credibility of drug–target associations.
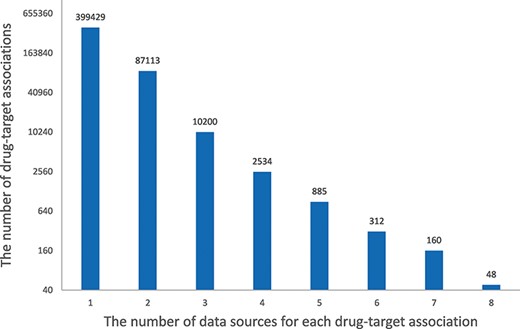
Histogram of the number of data sources for each drug–target association. The x-axis indicates the number of data sources for each drug–target association, and the y-axis indicates the number of drug–target associations.
Discussion
Drug–target association plays more and more roles in drug discovery, drug repositioning, drug synergy prediction, etc. HCDT database (http://hainmu-biobigdata.com/hcdt) is a publicly available resource for highly confident drug–target associations. The researchers can query drug–target associations via drugs or target genes and can download all the associations. HCDT database integrated eight databases and includes 500 681 highly confident (experimental verified) drug–target associations. It has the largest volume and has as much information as possible for drugs and target genes. Together with multi-omics data, it will be a good resource in analyzing the drug functional mechanism, mining drug-related biological pathway, predicting drug synergy, etc.
Supplementary data
Supplementary data are available at Database Online.
Acknowledgements
The authors thank all the anonymous referees for their valuable suggestions and support.
Funding
Major Science and Technology Program of Hainan Province (No. ZDKJ202003), National Natural Science Foundation of China (No. 32260155, 31701159 and 32160179), the Natural Science Foundation of Hainan Province (No. 621MS041, 821MS045, 821MS0777 and 822MS074); the Education Department of Hainan Province [No.Hnky2022-32]; the Innovation Fund for Postgraduates of Hainan Medical University (HYYS2021B12).
Conflict of interest
The authors declare no conflict of interest.
References
Author notes
have contributed equally to this work and share first authorship.



