-
PDF
- Split View
-
Views
-
Cite
Cite
Özge Kart, Alexandre Mestiashvili, Kurt Lachmann, Richard Kwasnicki, Michael Schroeder, Emati: a recommender system for biomedical literature based on supervised learning, Database, Volume 2022, 2022, baac104, https://doi.org/10.1093/database/baac104
Close - Share Icon Share
Abstract
The scientific literature continues to grow at an ever-increasing rate. Considering that thousands of new articles are published every week, it is obvious how challenging it is to keep up with newly published literature on a regular basis. Using a recommender system that improves the user experience in the online environment can be a solution to this problem. In the present study, we aimed to develop a web-based article recommender service, called Emati. Since the data are text-based by nature and we wanted our system to be independent of the number of users, a content-based approach has been adopted in this study. A supervised machine learning model has been proposed to generate article recommendations. Two different supervised learning approaches, namely the naïve Bayes model with Term Frequency-Inverse Document Frequency (TF-IDF) vectorizer and the state-of-the-art language model bidirectional encoder representations from transformers (BERT), have been implemented. In the first one, a list of documents is converted into TF-IDF–weighted features and fed into a classifier to distinguish relevant articles from irrelevant ones. Multinomial naïve Bayes algorithm is used as a classifier since, along with the class label, it also gives the probability that the input belongs to this class. The second approach is based on fine-tuning the pretrained state-of-the-art language model BERT for the text classification task. Emati provides a weekly updated list of article recommendations and presents it to the user, sorted by probability scores. New article recommendations are also sent to users’ email addresses on a weekly basis. Additionally, Emati has a personalized search feature to search online services’ (such as PubMed and arXiv) content and have the results sorted by the user’s classifier.
Database URL: https://emati.biotec.tu-dresden.de
Introduction
The scientific literature is growing rapidly. Since more and more articles are published every year, it can be hard to keep track of relevant topics. In 2020 alone, PubMed (https://pubmed.ncbi.nlm.nih.gov/) has indexed 1.4 million new scientific papers. This equals an average of 27 000 papers every week. With such high numbers, it is impossible to find the most relevant publication without tediously browsing through long pages of search results. Considering that a person can read an average of 200–250 words per minute and assuming that an average-length research article consists of 5000 words, the reading time of an article is a minimum of 20 min. According to this calculation, a researcher can read up to 168 articles per week by reading 8 h a day. These numbers show the difficulty of following the relevant literature for researchers with busy work routines. Recommendation systems play an important role in reducing this limitation and improving the search results by considering user profiles as well as domain data.
Recommender systems can be categorized into three approaches: collaborative filtering, content-based filtering and a hybrid approach. The content-based filtering approach considers the content similarity between users’ interests and the metadata of the articles (1, 2). Collaborative filtering offers suggestions based on the neighbor’s selection of the same user group (3). Lastly, the hybrid filtering technique aims to enhance recommendation quality by combining the first two methods (4). Content-based filtering is preferred when an item is information-rich, such as text data.
In a content-based recommendation system, a profile is created for each item based on the information it contains, and each user has a profile based on the items they have liked or disliked in the past. Thus, a user profile defines the type of content related to that user. The purpose of the system is to find items whose content best matches the data stored in this user profile.
Since the approach is domain-specific, this means that it can deliver more precise results. It is easier to accurately tell a user’s interests by looking at the actual content, rather than inferring from a group of similar users. Furthermore, the major advantage of a content-based design is that it is independent of the size of the user base. Recommendations will be equally accurate whether it is just a single user or millions of users. Therefore, it can be applied even to small projects that might not have enough users to make a collaborative approach applicable. However, in this approach, it is important to properly represent information in terms of features. In previous studies, articles are represented in different ways such as paper’s metadata (5), bag of words (6), vector space (7), key phrase (8, 9) and user’s given tag (10, 11). The TF-IDF method has been used as a similarity measure between items in article recommendation system studies (12, 13). There are many other studies (14–16) that use this method for the same purpose in different domains.
Recently, various approaches have been used in content-based research article recommendation systems. Chaudhuri et al. proposed new features for improving the efficiency of the recommendation system, which are extracted indirectly from research articles. These features are keyword diversification, text complexity, citation analysis over time and scientific quality measurement to represent a research article. The keyword diversification indicates the uniqueness of the keywords to help variation in a recommendation. The text complexity measure helps match an article to a user based on the user’s understandability level. The citation analysis over time indicates the relevancy of a paper. The scientific quality ranks the scientific merits of articles (17). Bulut et al. used the Doc2vec method to compare user profiles with candidate articles to be recommended. User profiles and candidate articles are represented as continuous vectors in the Doc2vec method. Similar articles to a user profile are recommended to that user (13). Hao et al. integrated academic network information with content similarity information to improve the accuracy and efficiency of recommendations. The academic network consists of links between author nodes, paper notes and conference nodes (18).
In content-based recommendation systems, the task of determining the semantic similarity of texts is challenging. The recently announced pretrained transformers for language modeling such as bidirectional encoder representations from transformers (BERT) Embeddings from Language Models and Generative Pre-trained Transformer 3 are quite successful in overcoming this challenge. These pretrained machine learning models have achieved state-of-the-art performance for many natural language processing (NLP) tasks such as question answering, sentiment analysis and named entity recognition (19–21). There are also some recent studies using BERT in various ways in recommendation systems. Jeong et al. (2020) proposed a citation recommender model by using BERT to obtain embedding vectors from textual data and by using graph convolutional network to obtain embedding vectors from citation graphs (22). Sun et al. proposed a recommender system in which BERT architecture was adapted to take a set of items instead of text as input values (23). In these studies, the BERT model showed a stronger performance than the other models.
According to a literature survey on research-paper recommender systems conducted by Beel et al. (24), out of the reviewed approaches, only 39% of research-paper recommender systems could be used by users in practice. Of these recommender systems, 44% are still running and actively maintained. On the other hand, Beel et al. emphasized that most of the real-world recommender systems implement basic recommendation approaches that are not based on recent research (24). In order to bridge this gap, we focused our study on developing a web-based article recommendation service, which provides users with updated research results.
The developed system adopts a content-based approach and uses supervised machine learning to create recommendations. The classification task is to label each article with one of the two target classes ‘interesting’ and ‘irrelevant’. Two different approaches have been implemented for this task. In the first one, we represent articles with TF-IDF vectors and train a classifier with these vectors. The multinomial naïve Bayes classifier is used in the implementation since it provides probability distribution, which determines the probability that the input belongs to each class, as well as a class label. The recommended articles are ranked based on their probability scores. The second approach is based on a state-of-the-art language model BERT. The pretrained BERT model is fine-tuned for the text classification task. The probability of class membership for each class label is obtained by the softmax function.
SCI-BERT is a pretrained language model that has the same architecture as BERT but is trained on a large corpus of scientific text (25). The SCI-BERT paper reports a +1.92 F1 score increase on average at the fine-tuned SCI-BERT models for various tasks in the biomedical domain such as named entity recognition, PICO extraction, relation classification and dependency parsing, compared to the BERT-base. However, it does not report any result for the text classification task in the biomedical domain, which we employ in this study. It reports the results of the text classification task on two data sets from multiple domains, which yield only a +0.49 F1 score increase on average. Therefore, large improvements are not expected from the use of SCI-BERT models instead of the BERT-base.
A website has been developed to display a weekly updated list of articles recommended for each user, which is available at https://emati.biotec.tu-dresden.de. The system also sends new article recommendations to users’ email addresses on a weekly basis. Furthermore, Emati has a personalized search feature to search from online services (such as PubMed and arXiv) and have the results sorted by the user’s classifier. The first approach, which is based on TF-IDF and multinomial naïve Bayes, is used in production as a classifier due to the need for scalability. The source codes of the project are available on the GitHub repository (https://github.com/bioinfcollab/emati), where both naïve Bayes– and BERT-based approaches are available to the user as a classifier option.
The advantages of Emati over similar systems such as arXivDigest (26) can be listed as follows: arXivDigest provides recommendations over papers published on only arXiv. Emati currently provides recommendations from both PubMed and arXiv, which means it can offer a wider range and produce more accurate recommendations, especially for biomedical researchers. Also due to the flexible design of Emati, further sources can be easily integrated. Moreover, arXivDigest requires the users to provide topics of their interests as keywords at the registration and it basically returns the keyword search results to the users as recommended articles. It enables the researchers can register their own recommender system; however, it does not itself provide any complex recommender engine based on machine learning, collaborative filtering or another state-of-the-art recommendation method. Emati recommendations are based on a machine learning classifier, and users can upload the reference list of the articles they are interested in to train the personalized classifier based on their interests.
Materials and Methods
Emati is a web-based recommender system that displays a weekly updated list of recommended scientific articles. Figure 1 shows a screenshot of the main page. The Emati database (DB) contains article information published in PubMed and arXiv since 2000, and it is updated every week with newly published articles. The approximate number of articles in the DB in 2022 is ∼15 million. The system allows fetching new content from any source, as long as it offers an Application Programming Interface for downloading papers with their title, abstract, journal, authors and date of publication stored in separate fields. Thus, it is possible to include multiple sources and expand the range of topics.
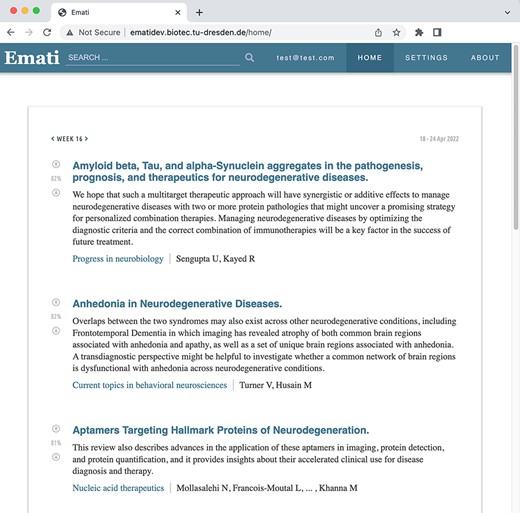
Weekly article recommendations of the user are listed on the homepage.
In order to get updated article recommendations, a user creates an account, logs in to the system and uploads their reference files. The file upload process is detailed in the 2.2.4 training corpus subsection. The reference files are parsed to learn about the topics this particular user is interested in. Once the initial recommender model is initialized, it can be updated with the data collected from user interaction. Users click on the articles to read and click like or dislike buttons in accordance with their interests. With this information, a personal machine learning classifier is trained. When enough new interactions are detected, the classifier is retrained to keep itself up-to-date. The trained model is used to create new weekly content for the user. A combination of percentage and absolute interactions is used as the number of new interactions required to trigger retraining. Whatever case occurs first triggers the retraining. The percentage threshold is the percentage in relation to total interactions so far. For example, a user has clicked/liked/disliked 100 articles in total. A threshold of 0.1 means that 100 * 0.1 = 10 new interactions since the last training are needed to train the classifier anew. The absolute threshold is the absolute number of new interactions required to trigger retraining. These parameters are set to 0.1 and 10 by default, respectively.
General structure of the system
The system is composed of two separate servers. Figure 2(a) shows the high-level system design of the project. The code implemented during this study resides within the Django project. The machine learning aspects of this project are also implemented in Python using the scikit-learn library.
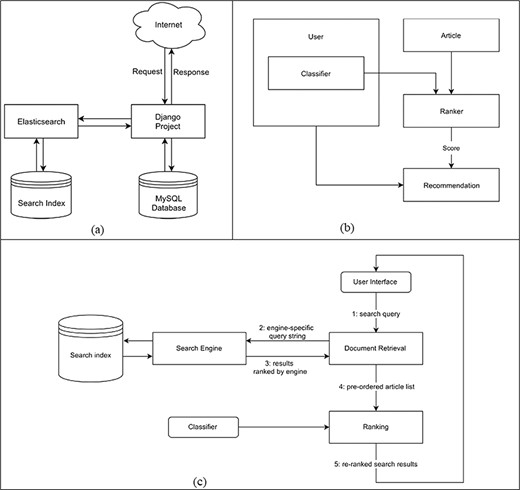
(a) High-level system design that shows how components of the system are connected and communicate with each other. (b) Data flow diagram for the creation of recommendations. (c) Personalized search pipeline that shows how a query term is searched from the search engine and returned results are reordered by the classifier.
Elasticsearch is the search engine responsible for the full-text search feature. Django communicates with the search index by sending requests to Elasticsearch’s web interface. Elasticsearch consists of a search index managed by a server that provides an HTTP web interface. By sending requests to that server, it is possible to query the search index or add new documents. There is also an official Python client for Elasticsearch that takes care of handling requests and responses (https://github.com/elastic/elasticsearch-py, accessed 30 October 2021). This makes it very easy to communicate with the search index from within the Django project.
Recommender model
The recommender model is based on a classification task that is used to label each article with one of the two target classes ‘interesting’ and ‘irrelevant’. Since our ultimate goal is to rank articles according to how interesting they are, we need not only a binary classification but also their probability of belonging to a particular target class. This requirement was met by using the multinomial naïve Bayes classifier and the BERT classifier with a softmax function in the implementation of this study. Since those classes are mutually exclusive, their scores will always add up to 1. In other words, if an article is considered interesting with a probability of 80%, then it must be irrelevant with a probability of 20%.
Figure 2(b) shows the data flow during the creation of new recommendations. The Ranker will take a list of articles together with the classifier of a single user. It then calculates the scores for each article. These scores indicate the probability with which an article belongs to the class titled interesting. For the best scoring articles, the Ranker will create Recommendation objects that contain references to the user and that specific article together with the computed score. By saving only the best scoring recommendations, we also reduce the required data storage.
Multinomial naïve Bayes
where t is the target feature with a set of levels, levels(t), and q is the query instance with a set of descriptive features, q[1], …, q[m].
Feature generation and classification
Our data set consists of the article’s title and abstract, the journal it was published in, the list of authors and the date of publication. These data can be used to categorize a document into ‘interesting’ and ‘irrelevant’. The text is split into words that are then weighted according to the TF-IDF weighting scheme. Each document is represented by a vector of these features. The vectors are then fed into the multinomial naïve Bayes classifier.
During training, the algorithm will learn all words present in the corpus. Since each user’s field of interest is different, each training corpus will be different as well. This also means that all users will have a personalized vocabulary of words that are known to their classifier. Each user’s final model, therefore, consists of a vectorizer and a classifier. The vectorizer represents the learned vocabulary. It is responsible for converting a list of documents into vectors of TF-IDF–weighted features. During the training process, the vectorizer learns the weights based on the given corpus. The TF-IDF vectorizer uses the corpus from the training data. So while creating recommendations from new articles, it can vectorize only existing terms in that corpus. The classifier is responsible for the actual categorization task. It takes a list of feature vectors and returns a vector of probabilities for each document, which indicates the likeliness with which it belongs to one of the available classes.
Bidirectional encoder representations from transformers
BERT is one of the most popular natural language models which is mainly divided into two stages: pretraining and fine-tuning. It has been pretrained to learn deep bidirectional representation from the unlabeled text. It is designed to perform fine-tuning using labeled text for various NLP tasks after pretraining (19). The BERT model uses a large corpus comprising BooksCorpus (800 M words) (28) and English Wikipedia (2500 M words) for the pretraining. In fine-tuning, the BERT model is initialized with pretrained parameters and then trained on a downstream task, which is text classification in our case, by simply fine-tuning all pretrained parameters. The self-attention mechanism in the transformer enables the modeling of multiple downstream tasks by replacing appropriate inputs and outputs.
BERT’s most superior feature is to generate contextualized word representations. However, it also brings some drawbacks. The model is huge due to the training structure and large corpus. It is slower to train compared to shallow machine learning algorithms. It is also very compute-intensive at inference time, which means that it can become costly if someone wants to use it in production at scale. A single BERT-base model checkpoint is ∼1.3 GB in size. Moreover, it is stated that the fine-tuning examples in the BERT paper (19), which use BERT-base, should be able to run on a graphics processing unit that has at least 12 GB of random access memory (RAM) using the hyperparameters given in https://github.com/google-research/bert. In a multiuser system where each user’s own profile is created and updated over time, such high-capacity hardware requirements increase costs significantly as the number of users increases.
Training corpus
As already stated earlier, the recommender system works on scientific articles. But naturally, the system has no data on a newly signed-up user. To overcome this problem, which is also referred to as the ‘cold-start’ or ‘new-user’ problem, the system provides users with the possibility to upload reference files from the ‘Settings’ menu, as shown in Figure 3. Currently supported file formats are BibTeX (.bib), RIS (.ris) and Endnote XML (.xml). These files contain a list of articles the user has cited in the past. This makes it possible to infer the field they are working on. The reference files contain the same type of data that our system is working with (title, journal, author and abstract) and already store it in separate fields. Thus, they can be easily parsed and fed into the algorithm. If the abstract of an article is not already provided in the reference file, the system queries the article title from the sources and includes the abstract if available. Users can also upload a PubMed ID (PMID) list of articles as a text (.txt) file or save the PMIDs in a text area provided in the ‘Settings’ menu. The system queries PubMed by the provided PMIDs and fetches the article information such as title, journal, author and abstract. According to a current study (29), the increase of the Area Under the Curve value when full texts were used instead of abstracts ranges from 0% to 9% across six different text mining tasks, and the median was 3.5%. Since the improvement expected in the performance is not so large compared to the reduced scalability and increase in resources needed, we decided to use abstracts in this study.
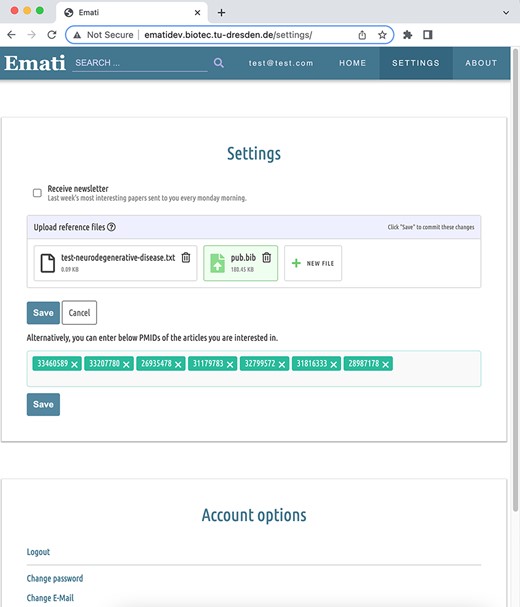
The data points collected this way serve as positive training samples—they define the ‘interesting’ class. A set of randomly picked articles are used as negative samples. The training corpus is filled with random articles until there is an equal amount of positive and negative samples. Once the initial recommender model is initialized, it can be updated with the data collected from user interaction. Every user interaction is logged so that a classifier can be retrained regularly with an ever-increasing corpus. Implicitly, this is done by incorporating the articles that the user clicked on to view their details since it might be an indication of their interest. More explicit feedback is provided by the website in the form of like and dislike buttons. The collected feedback is weighted higher than the logged click since the user explicitly tells the system their opinion. In other words, clicked articles have less (50% by default) weight than liked articles while training a classifier since ‘like’ is stronger evidence rather than ‘click’. Also with disliked articles, a better negative training set is obtained. During the first training, the negative samples consisted only of randomly picked articles, meaning that the system tried to discern ‘interesting’ articles from the ‘average’ article. But it cannot make a clear distinction since it does not know the true bounds of the ‘interesting’ class—the borders are fuzzy. By using articles that were considered ‘interesting’ by the engine but labeled ‘irrelevant’ by the user, the true edge of the ‘interesting’ class can be discovered and the border becomes sharper.
Results and Discussion
In order to evaluate the proposed recommendation system, we conducted some necessary experiments on several data sets for different use-case scenarios. All the experiments were implemented using Python on a Debian GNU/Linux 10 (buster) and run on an Intel(R) Xeon(R) E5-2650 v2 @ 2.60 GHz central processing unit and 256 GB RAM. Both the naïve Bayes with TF-IDF vectorizer and BERT-based approaches have been implemented. The multinomial naïve Bayes algorithm was implemented using scikit-learn. The pretrained ‘BERT-base-uncased’ model has been downloaded and fine-tuned for the text classification task using the HuggingFace Transformers library on the data sets detailed later. The text data are tokenized by using the BERT tokenizer and fed to the BERT model. The input token length is 300, which is the approximate length of an abstract. The parameters set for fine-tuning are as follows: the number of training epochs is 3. Batch sizes for training and evaluation are set to 32 and 64, respectively. The best model is loaded to be used for inference at the end of training.
A 10-fold cross-validation scheme has been implemented to evaluate the naïve Bayes models. n-fold cross-validation splits the training data into n equal parts. A model is trained by n − 1 parts and tested by one part. This process iterates n times until all parts are used in the training and test process. To be more specific, the split ratio is 90:10 per iteration in our case. The hold-out validation method has been used to evaluate the BERT models. The split ratio is 80:20.
A researcher use case with a particular research focus
A new-user account has been created, and a BibTeX file containing positive training examples has been generated and uploaded to Emati. In order to create a user profile for a researcher who is interested in biology with a specific focus on liquid–liquid phase separation (LLPS), 52 protein names that are common in four LLPS DBs (30–33) have been queried in PubMed along with related keywords such as phase separation, phase transition, condensate and membraneless organelle. The articles published in journals with an impact factor have been filtered out in the results to create a positive data set. As a result, the reference file contained data of 300 articles. The same number of negative samples has been randomly selected among the downloaded PubMed articles published between 1 May 2021 and 10 July 2021. Two more example user profiles have been created separately for the researchers who are interested in the topics of ‘neurodegenerative diseases’ and ‘antibiotics resistance’. These two terms have been queried in PubMed. The articles published in journals with an impact factor have been filtered out in the results to create positive data sets from indexed journals. As a result, the data of 7048 and 6852 articles have been uploaded to the system as positive samples for example user profiles who are interested in the topics of neurodegenerative diseases and antibiotics resistance, respectively. The same number of negative samples has been randomly selected among the downloaded PubMed articles published in 2021.
The performance values obtained by the experiments using both the naïve Bayes and BERT models are given in Tables 1 and 2. BERT models provided more accurate results compared to naïve Bayes models for all researcher profiles. We also have subsampled 15 random samples from each constructed positive data set and trained models using them to observe the training performance of the models when users provide a small set of positive examples. When using smaller training sets, the performance drop of BERT models was quite greater than that of naïve Bayes. The naïve Bayes performed better on small data sets.
Training performance values obtained by the validation of naïve Bayes models trained with three different researcher profiles
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 0.95 | 0.95 | 0.83 | 0.7 | 0.88 | 0.65 |
| Recall | 1 | 0.9 | 0.96 | 1 | 0.99 | 1 |
| F-measure | 0.98 | 0.95 | 0.89 | 0.82 | 0.93 | 0.79 |
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 0.95 | 0.95 | 0.83 | 0.7 | 0.88 | 0.65 |
| Recall | 1 | 0.9 | 0.96 | 1 | 0.99 | 1 |
| F-measure | 0.98 | 0.95 | 0.89 | 0.82 | 0.93 | 0.79 |
Training performance values obtained by the validation of naïve Bayes models trained with three different researcher profiles
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 0.95 | 0.95 | 0.83 | 0.7 | 0.88 | 0.65 |
| Recall | 1 | 0.9 | 0.96 | 1 | 0.99 | 1 |
| F-measure | 0.98 | 0.95 | 0.89 | 0.82 | 0.93 | 0.79 |
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 0.95 | 0.95 | 0.83 | 0.7 | 0.88 | 0.65 |
| Recall | 1 | 0.9 | 0.96 | 1 | 0.99 | 1 |
| F-measure | 0.98 | 0.95 | 0.89 | 0.82 | 0.93 | 0.79 |
Training performance values obtained by the validation of BERT models trained with three different researcher profiles
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 1 | 0.5 | 0.97 | 0.33 | 0.98 | 0.66 |
| Recall | 0.98 | 1 | 0.97 | 1 | 0.98 | 1 |
| F-measure | 0.99 | 0.67 | 0.99 | 0.5 | 0.98 | 0.8 |
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 1 | 0.5 | 0.97 | 0.33 | 0.98 | 0.66 |
| Recall | 0.98 | 1 | 0.97 | 1 | 0.98 | 1 |
| F-measure | 0.99 | 0.67 | 0.99 | 0.5 | 0.98 | 0.8 |
Training performance values obtained by the validation of BERT models trained with three different researcher profiles
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 1 | 0.5 | 0.97 | 0.33 | 0.98 | 0.66 |
| Recall | 0.98 | 1 | 0.97 | 1 | 0.98 | 1 |
| F-measure | 0.99 | 0.67 | 0.99 | 0.5 | 0.98 | 0.8 |
| Researcher profile . | LLPS proteins . | Neurodegenerative diseases . | Antibiotics resistance . | |||
|---|---|---|---|---|---|---|
| Data set size | Large (300 * 2) | Small (15 * 2) | Large (7048 * 2) | Small (15 * 2) | Large (6852 * 2) | Small (15 * 2) |
| Precision | 1 | 0.5 | 0.97 | 0.33 | 0.98 | 0.66 |
| Recall | 0.98 | 1 | 0.97 | 1 | 0.98 | 1 |
| F-measure | 0.99 | 0.67 | 0.99 | 0.5 | 0.98 | 0.8 |
A curator use case with a focus on a model organism DB
Model organism DBs are biological DBs that holistically produce, source and blend species-specific information by putting expert knowledge together with bioinformatics and literature curation. DB curators are hired experts who read articles, extract useful information and place that information into searchable DBs.
Considering that the curators may be potential users of the proposed system, some retrospective experiments have been carried out on FlyBase (http://flybase.org/), The Zebrafish Information Network (ZFIN) (http://zfin.org/) and Mouse Genome Informatics (MGI) (http://www.informatics.jax.org/), which are model organism DBs for fruit fly, zebrafish and mouse, respectively. The DBs provide the information of the articles they use as evidence for their annotations. In our experiments, we have extracted those articles published in 2019 to be used as a positive training set while training recommender models. The number of articles extracted from each DB is given in Table 3. The same number of random articles published in 2019 has been collected from PubMed as negative examples. Table 4 shows the precision, recall and F-measure values of the trained models for each DB based on the implemented validation scheme.
| DB name . | FlyBase . | ZFIN . | MGI . |
|---|---|---|---|
| No. of articles in 2019 cited in the DB | 2825 | 3330 | 12 744 |
| DB name . | FlyBase . | ZFIN . | MGI . |
|---|---|---|---|
| No. of articles in 2019 cited in the DB | 2825 | 3330 | 12 744 |
| DB name . | FlyBase . | ZFIN . | MGI . |
|---|---|---|---|
| No. of articles in 2019 cited in the DB | 2825 | 3330 | 12 744 |
| DB name . | FlyBase . | ZFIN . | MGI . |
|---|---|---|---|
| No. of articles in 2019 cited in the DB | 2825 | 3330 | 12 744 |
Training performance values obtained by the validation of naïve Bayes and BERT classifiers trained with the FlyBase, ZFIN and MGI data sets
| . | Naïve Bayes (10-fold cv) . | BERT (train/test split 80:20) . | ||||
|---|---|---|---|---|---|---|
| DB name | FlyBase | ZFIN | MGI | FlyBase | ZFIN | MGI |
| Precision | 0.97 | 0.97 | 0.98 | 0.98 | 0.97 | 0.94 |
| Recall | 0.82 | 0.75 | 0.85 | 0.97 | 0.94 | 0.98 |
| F-measure | 0.89 | 0.84 | 0.91 | 0.97 | 0.95 | 0.96 |
| . | Naïve Bayes (10-fold cv) . | BERT (train/test split 80:20) . | ||||
|---|---|---|---|---|---|---|
| DB name | FlyBase | ZFIN | MGI | FlyBase | ZFIN | MGI |
| Precision | 0.97 | 0.97 | 0.98 | 0.98 | 0.97 | 0.94 |
| Recall | 0.82 | 0.75 | 0.85 | 0.97 | 0.94 | 0.98 |
| F-measure | 0.89 | 0.84 | 0.91 | 0.97 | 0.95 | 0.96 |
Training performance values obtained by the validation of naïve Bayes and BERT classifiers trained with the FlyBase, ZFIN and MGI data sets
| . | Naïve Bayes (10-fold cv) . | BERT (train/test split 80:20) . | ||||
|---|---|---|---|---|---|---|
| DB name | FlyBase | ZFIN | MGI | FlyBase | ZFIN | MGI |
| Precision | 0.97 | 0.97 | 0.98 | 0.98 | 0.97 | 0.94 |
| Recall | 0.82 | 0.75 | 0.85 | 0.97 | 0.94 | 0.98 |
| F-measure | 0.89 | 0.84 | 0.91 | 0.97 | 0.95 | 0.96 |
| . | Naïve Bayes (10-fold cv) . | BERT (train/test split 80:20) . | ||||
|---|---|---|---|---|---|---|
| DB name | FlyBase | ZFIN | MGI | FlyBase | ZFIN | MGI |
| Precision | 0.97 | 0.97 | 0.98 | 0.98 | 0.97 | 0.94 |
| Recall | 0.82 | 0.75 | 0.85 | 0.97 | 0.94 | 0.98 |
| F-measure | 0.89 | 0.84 | 0.91 | 0.97 | 0.95 | 0.96 |
The models trained on the full 2019 training data have been used to create recommendations for the first, second and third weeks of 2020. The system recommendations have been compared with the articles published in corresponding weeks which were already cited by the DBs. Precision@50 (precision at top 50 Emati recommendations) and Recall@n for different n values have been computed for each DB (Tables 5–7).
Recommendation performance values of the classifiers trained with the FlyBase data set, based on the first 3 weeks of 2020
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in FlyBase) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 58 | 11/58 = 0.19 | 11/50 = 0.22 | 33/58 = 0.57 | 29/50 = 0.58 |
| Second week of 2020 | 66 | 23/66 = 0.35 | 17/50 = 0.34 | 48/66 = 0.73 | 39/50 = 0.78 |
| Third week of 2020 | 46 | 15/46 = 0.33 | 17/50 = 0.34 | 26/46 = 0.57 | 27/50 = 0.54 |
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in FlyBase) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 58 | 11/58 = 0.19 | 11/50 = 0.22 | 33/58 = 0.57 | 29/50 = 0.58 |
| Second week of 2020 | 66 | 23/66 = 0.35 | 17/50 = 0.34 | 48/66 = 0.73 | 39/50 = 0.78 |
| Third week of 2020 | 46 | 15/46 = 0.33 | 17/50 = 0.34 | 26/46 = 0.57 | 27/50 = 0.54 |
Recommendation performance values of the classifiers trained with the FlyBase data set, based on the first 3 weeks of 2020
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in FlyBase) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 58 | 11/58 = 0.19 | 11/50 = 0.22 | 33/58 = 0.57 | 29/50 = 0.58 |
| Second week of 2020 | 66 | 23/66 = 0.35 | 17/50 = 0.34 | 48/66 = 0.73 | 39/50 = 0.78 |
| Third week of 2020 | 46 | 15/46 = 0.33 | 17/50 = 0.34 | 26/46 = 0.57 | 27/50 = 0.54 |
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in FlyBase) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 58 | 11/58 = 0.19 | 11/50 = 0.22 | 33/58 = 0.57 | 29/50 = 0.58 |
| Second week of 2020 | 66 | 23/66 = 0.35 | 17/50 = 0.34 | 48/66 = 0.73 | 39/50 = 0.78 |
| Third week of 2020 | 46 | 15/46 = 0.33 | 17/50 = 0.34 | 26/46 = 0.57 | 27/50 = 0.54 |
Recommendation performance values of the classifiers trained with the ZFIN data set, based on the first 3 weeks of 2020
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in ZFIN) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 178 | 63/178 = 0.35 | 33/50 = 0.66 | 108/178 = 0.61 | 48/50 = 0.96 |
| Second week of 2020 | 38 | 5/38 = 0.13 | 9/50 = 0.18 | 28/38 = 0.74 | 31/50 = 0.62 |
| Third week of 2020 | 58 | 12/58 = 0.21 | 11/50 = 0.22 | 42/58 = 0.72 | 42/50 = 0.84 |
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in ZFIN) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 178 | 63/178 = 0.35 | 33/50 = 0.66 | 108/178 = 0.61 | 48/50 = 0.96 |
| Second week of 2020 | 38 | 5/38 = 0.13 | 9/50 = 0.18 | 28/38 = 0.74 | 31/50 = 0.62 |
| Third week of 2020 | 58 | 12/58 = 0.21 | 11/50 = 0.22 | 42/58 = 0.72 | 42/50 = 0.84 |
Recommendation performance values of the classifiers trained with the ZFIN data set, based on the first 3 weeks of 2020
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in ZFIN) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 178 | 63/178 = 0.35 | 33/50 = 0.66 | 108/178 = 0.61 | 48/50 = 0.96 |
| Second week of 2020 | 38 | 5/38 = 0.13 | 9/50 = 0.18 | 28/38 = 0.74 | 31/50 = 0.62 |
| Third week of 2020 | 58 | 12/58 = 0.21 | 11/50 = 0.22 | 42/58 = 0.72 | 42/50 = 0.84 |
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in ZFIN) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 178 | 63/178 = 0.35 | 33/50 = 0.66 | 108/178 = 0.61 | 48/50 = 0.96 |
| Second week of 2020 | 38 | 5/38 = 0.13 | 9/50 = 0.18 | 28/38 = 0.74 | 31/50 = 0.62 |
| Third week of 2020 | 58 | 12/58 = 0.21 | 11/50 = 0.22 | 42/58 = 0.72 | 42/50 = 0.84 |
Recommendation performance values of the classifiers trained with the MGI data set, based on the first 3 weeks of 2020
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in MGI) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 505 | 102/505a = 0.20 | 27/50 = 0.54 | 170/505 = 0.34 | 37/50 = 0.74 |
| Second week of 2020 | 151 | 52/151 = 0.34 | 20/50 = 0.40 | 49/151 = 0.32 | 30/50 = 0.60 |
| Third week of 2020 | 184 | 28/184 = 0.15 | 19/50 = 0.38 | 63/184 = 0.34 | 31/50 = 0.62 |
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in MGI) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 505 | 102/505a = 0.20 | 27/50 = 0.54 | 170/505 = 0.34 | 37/50 = 0.74 |
| Second week of 2020 | 151 | 52/151 = 0.34 | 20/50 = 0.40 | 49/151 = 0.32 | 30/50 = 0.60 |
| Third week of 2020 | 184 | 28/184 = 0.15 | 19/50 = 0.38 | 63/184 = 0.34 | 31/50 = 0.62 |
The number of weekly Emati recommendations < n.
Recommendation performance values of the classifiers trained with the MGI data set, based on the first 3 weeks of 2020
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in MGI) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 505 | 102/505a = 0.20 | 27/50 = 0.54 | 170/505 = 0.34 | 37/50 = 0.74 |
| Second week of 2020 | 151 | 52/151 = 0.34 | 20/50 = 0.40 | 49/151 = 0.32 | 30/50 = 0.60 |
| Third week of 2020 | 184 | 28/184 = 0.15 | 19/50 = 0.38 | 63/184 = 0.34 | 31/50 = 0.62 |
| . | . | Naïve Bayes classifier . | BERT classifier . | ||
|---|---|---|---|---|---|
| n (no. of weekly articles in MGI) | Recall@n | Precision@50 | Recall@n | Precision@50 | |
| First week of 2020 | 505 | 102/505a = 0.20 | 27/50 = 0.54 | 170/505 = 0.34 | 37/50 = 0.74 |
| Second week of 2020 | 151 | 52/151 = 0.34 | 20/50 = 0.40 | 49/151 = 0.32 | 30/50 = 0.60 |
| Third week of 2020 | 184 | 28/184 = 0.15 | 19/50 = 0.38 | 63/184 = 0.34 | 31/50 = 0.62 |
The number of weekly Emati recommendations < n.
The training performance of the naïve Bayes classifiers trained using the same number of positive and negative examples is quite high with F-measures ranging from 0.84 to 0.91. However, since thousands of new papers are published in a week, the task of recommending weekly articles is much harder than distinguishing positive and negative examples in a balanced data set. Therefore, it is reasonable that the precision and recall values given in Tables 5–7 are relatively lower. On the other hand, the BERT models obtained outstanding results for the recommendation task. The recommendation performance values of fine-tuned BERT models are significantly higher than naïve Bayes models. Figure 4 illustrates the 3-week average recommendation performance of classifiers trained with the FlyBase, ZFIN and MGI data sets, whose sizes are given next to their names.
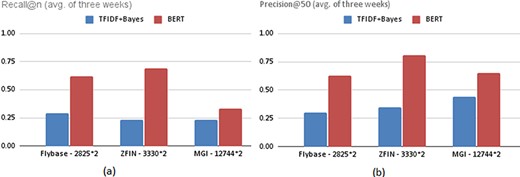
Average recommendation performances of the classifiers trained with the FlyBase, ZFIN and MGI data sets in terms of (a) Recall@n and (b) Precision@50.
Model training time and size evaluation
The training time of each model trained on the smallest (LLPS) and largest (MGI) data sets used in the experiments is given in Table 8. The size of the data set and the complexity of the model are highly related to the training time. The sizes of BERT models are huge due to the training structure and large corpus compared to the naïve Bayes models, as shown in Table 8. The naïve Bayes model sizes include both vectorizer and classifier.
Training time and size of the naïve Bayes and BERT models trained on the LLPS and MGI data sets
| . | . | Model name . | |||
|---|---|---|---|---|---|
| . | . | Naïve Bayes . | BERT . | ||
| Data set name | Data set size | Training time | Model size | Training time | Model size |
| LLPS | 300 * 2 | 0.32 s | 1.1 MB | 18 min 45 s | 1.3 GB |
| MGI | 12 744 * 2 | 9.34 s | 13 MB | 12 h 56 min 12 s | 1.3 GB |
| . | . | Model name . | |||
|---|---|---|---|---|---|
| . | . | Naïve Bayes . | BERT . | ||
| Data set name | Data set size | Training time | Model size | Training time | Model size |
| LLPS | 300 * 2 | 0.32 s | 1.1 MB | 18 min 45 s | 1.3 GB |
| MGI | 12 744 * 2 | 9.34 s | 13 MB | 12 h 56 min 12 s | 1.3 GB |
Training time and size of the naïve Bayes and BERT models trained on the LLPS and MGI data sets
| . | . | Model name . | |||
|---|---|---|---|---|---|
| . | . | Naïve Bayes . | BERT . | ||
| Data set name | Data set size | Training time | Model size | Training time | Model size |
| LLPS | 300 * 2 | 0.32 s | 1.1 MB | 18 min 45 s | 1.3 GB |
| MGI | 12 744 * 2 | 9.34 s | 13 MB | 12 h 56 min 12 s | 1.3 GB |
| . | . | Model name . | |||
|---|---|---|---|---|---|
| . | . | Naïve Bayes . | BERT . | ||
| Data set name | Data set size | Training time | Model size | Training time | Model size |
| LLPS | 300 * 2 | 0.32 s | 1.1 MB | 18 min 45 s | 1.3 GB |
| MGI | 12 744 * 2 | 9.34 s | 13 MB | 12 h 56 min 12 s | 1.3 GB |
Personalized search
The system provides a personalized search feature. Users can search articles’ content and have the results sorted according to their classifier. This allows for better search results because the system already knows which topics are relevant to the user without having to explicitly narrow down the search query. The search pipeline is illustrated in Figure 2(c). Instead of querying an online service (such as PubMed) for every search request, which slows down the search, the design is centered on a DB storing all fetched articles. Searching is done on a local search index. The index is updated regularly with new publications.
Conclusion and future work
We have developed a web-based article recommender service which can be accessed at https://emati.biotec.tu-dresden.de. It displays a weekly updated list of articles based on the users’ profile and sends it to users’ emails on a weekly basis. It has also a personalized search feature to search online services’ (such as PubMed and arXiv) content and have the results sorted by the user’s classifier. The potential users of the recommender system are scientific researchers who want to keep up-to-date with the scientific literature. For example, model organism DB curators who read the currently published relevant articles, extract useful information and place that information into searchable DBs are potential users to highly benefit from the system. In addition, the system also might be useful for PhD students, postdoctoral researchers or principal investigators who want to follow current literature related to their research interests, as well as research group members who want to present current articles in the journal club.
The system contains a content-based recommender using supervised machine learning to classify articles as ‘relevant’ and ‘irrelevant’ to the user. Two different supervised learning approaches, namely the naïve Bayes model with TF-IDF vectorizer and the state-of-the-art language model BERT, have been implemented. The training performance of naïve Bayes models trained on small data sets was higher than that of fine-tuned BERT models. On the other hand, BERT models showed outstanding recommendation performance in the experiments using test data sets from model organism DBs. Due to the need for scalability, the currently deployed version of Emati is based on the TF-IDF vectorizer and the naïve Bayes classifier approach. The BERT fine-tuning implementation is also available as an option in the source code which is provided at https://github.com/bioinfcollab/emati.
Currently, the system sources new content from PubMed and arXiv. In the future, it could still be extended by incorporating even more additional services as sources.
Acknowledgements
We are very grateful to Steven Marygold from FlyBase, Douglas G. Howe and Leyla Ruzicka from ZFIN and Martin Ringwald and Cynthia Smith from MGI for their valuable feedback on this study.
Funding
Federal Ministry of Education and Research project Center for Scalable Data Analytics and Artificial Intelligence.
Conflict of interest
None declared.



