-
PDF
- Split View
-
Views
-
Cite
Cite
Rezarta Islamaj, Chih-Hsuan Wei, Po-Ting Lai, Ling Luo, Cathleen Coss, Preeti Gokal Kochar, Nicholas Miliaras, Oleg Rodionov, Keiko Sekiya, Dorothy Trinh, Deborah Whitman, Zhiyong Lu, The biomedical relationship corpus of the BioRED track at the BioCreative VIII challenge and workshop, Database, Volume 2024, 2024, baae071, https://doi.org/10.1093/database/baae071
Close - Share Icon Share
Abstract
The automatic recognition of biomedical relationships is an important step in the semantic understanding of the information contained in the unstructured text of the published literature. The BioRED track at BioCreative VIII aimed to foster the development of such methods by providing the participants the BioRED-BC8 corpus, a collection of 1000 PubMed documents manually curated for diseases, gene/proteins, chemicals, cell lines, gene variants, and species, as well as pairwise relationships between them which are disease–gene, chemical–gene, disease–variant, gene–gene, chemical–disease, chemical–chemical, chemical–variant, and variant–variant. Furthermore, relationships are categorized into the following semantic categories: positive correlation, negative correlation, binding, conversion, drug interaction, comparison, cotreatment, and association. Unlike most of the previous publicly available corpora, all relationships are expressed at the document level as opposed to the sentence level, and as such, the entities are normalized to the corresponding concept identifiers of the standardized vocabularies, namely, diseases and chemicals are normalized to MeSH, genes (and proteins) to National Center for Biotechnology Information (NCBI) Gene, species to NCBI Taxonomy, cell lines to Cellosaurus, and gene/protein variants to Single Nucleotide Polymorphism Database. Finally, each annotated relationship is categorized as ‘novel’ depending on whether it is a novel finding or experimental verification in the publication it is expressed in. This distinction helps differentiate novel findings from other relationships in the same text that provides known facts and/or background knowledge. The BioRED-BC8 corpus uses the previous BioRED corpus of 600 PubMed articles as the training dataset and includes a set of newly published 400 articles to serve as the test data for the challenge. All test articles were manually annotated for the BioCreative VIII challenge by expert biocurators at the National Library of Medicine, using the original annotation guidelines, where each article is doubly annotated in a three-round annotation process until full agreement is reached between all curators. This manuscript details the characteristics of the BioRED-BC8 corpus as a critical resource for biomedical named entity recognition and relation extraction. Using this new resource, we have demonstrated advancements in biomedical text-mining algorithm development.
Database URL: https://codalab.lisn.upsaclay.fr/competitions/16381
Introduction
Biomedical entities appear throughout the biomedical research literature, in studies from chemistry, biology, and genetics to various other disciplines such as medicine and pharmacology. Naturally, the names of these biomedical entities are frequently searched in PubMed [1]. The development of automatic information retrieval and document understanding tools, therefore, necessitates the correct identification of biomedical entities mentioned in the text. This correct recognition of the mentioned or referenced entities has a significant impact on document retrieval, helping scientists search and find the relevant literature, as well as helping biocurators extract and catalogue this information into the relevant knowledge sources.
Biocurators are domain experts who can skillfully distil the information present in the unstructured text of a published paper into structured information that lists bioentities, their structure, their uses, and other characteristics and features that could be important for medicine, biology, and human health. Following this, as illustrated in Fig. 1, the National Library of Medicine (NLM) provides a suite of databases and resources essential for biomedical and health-related research: National Center for Biotechnology Information (NCBI) Gene [2] offers comprehensive information on genes, supporting genomic studies across a variety of organisms; NCBI Taxonomy [3] offers a detailed classification system for all organisms in genetic databases, improving data organization; Single Nucleotide Polymorphism Database (dbSNP) [4] is a vast repository of single-nucleotide polymorphisms and genetic variations, crucial for research on genetic variation; ClinVar [5] aggregates data on genomic variations, underlining their significance for human health and aiding in diagnostics; and MeSH [6] supplies a standardized vocabulary for efficient indexing, cataloguing, and retrieval of all the medical, health, and biomedical information. Furthermore, the Comparative Toxicogenomics Database [7] catalogues the interplay between chemical exposures, gene interactions, and disease outcomes, offering insight into the environmental impacts on health. The information curated in PharmGKB [8] connects genetic variations to drug responses and, in turn, facilitates research on personalized medicine. UniProt [9] is a vital resource for proteomics and molecular biology, providing detailed data on protein sequences and functions. Genome-wide association study [10] methodologies are pivotal in identifying genetic variants associated with diseases through extensive genome analysis. DisGeNET [11] aggregates gene–disease association data, enhancing research in genetics and the understanding of complex diseases. Together, these resources and others that we could not include in the image due to space constraints offer the meticulously curated and structured information covering the genetic and molecular foundations of health and disease, propelling research and applications in genomics, pharmacogenomics, proteomics, and disease understanding, smoothly integrating the introduction of these databases into the broader context of biomedical research support.
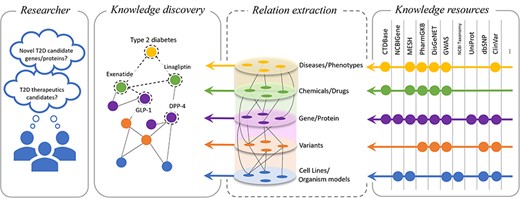
Overview of the relationship extraction in the biomedical domain. This illustration depicts the importance of factual and correct knowledge discovery based on verifiable facts curated in the knowledge bases.
How can the structured information catalogued in the knowledge resources propel research? Most research questions [e.g. ‘What are the T2D (Type 2 diabetes) therapeutics candidates?’] necessitate not only a correct automatic identification of these entities but also the understanding of their interactions, their usage, and their implications with respect to disease and human health (Fig. 1). This is particularly important in the era of large language models, where developing automated algorithms that correctly capture the semantic relationships contained in the text is crucially important to eliminate hallucinations and other spurious correlations, which may have significant problems in the medical and biomedical domains [12].
BioCreative challenges have consistently embraced the relation extraction tasks since 2007 and provided manually annotated corpora to support the development of automatic tools that can reliably perform these tasks. The corpus for BioCreative II Protein-Protein Interaction task [13] provided manual annotation for the curated protein interactions within a set of full-text journal articles. The developed tools focused on identifying the proteins mentioned in the article text and on extracting the pairs of proteins, the interactions of which were detailed in the articles. The BioCreative II.5 interaction pair extraction task [14] followed the previous challenge and furthermore identified the protein mentions with their corresponding UniProt identifiers. Thus, the developed tools needed to normalize the entity mentions in the article text to their identifiers and the interaction was expressed as a pair of protein identifiers. The BioCreative IV CTD [15] and BioCreative V chemical-disease relation [16] tasks introduced corpora that detailed relations between different types of entities, employing multiple standardized vocabularies. The tasks challenged participants to extract relations between chemicals, diseases, and genes, and these interactions were identified by the respective entities’ corresponding database identifiers (i.e. chemical–gene, disease–gene, and chemical–disease relations). The BioCreative VI precision medicine task [17] centered on precision medicine and its corpus consisted of triple relationships, in that the interaction (or the loss of interaction) between a given pair of proteins/genes was observed only in the presence (or absence) of a mutation. Genes and proteins were identified by their respective NCBI Gene record, and the tools used the context to predict whether the interaction was dependent on (caused by) a genetic mutation. The BioCreative VI ChemProt [18] and BioCreative VII DrugProt tasks [19] addressed chemical/drug–protein interactions in the context of drug discovery, and their corpus provided manual annotation for mentioned proteins, genes, and all binary relationships between them corresponding to a specific set of biologically relevant relation types such as ‘inhibitor’ and ‘activator’; however, no standardized vocabulary was used.
In addition, there are several other notable community challenges that have provided curated relation datasets in the biomedical domain. SemEval tasks [20–24] have included specific challenges focused on biomedical entities such as chemicals and diseases, and their biomedical relation extraction tasks such as drug–drug interaction [20] and temporal relation extraction task [23] have fostered developments in semantic analysis within biology and medicine. The Informatics for Integrating Biology and the Bedside [25] has provided a platform for developing and evaluating Natural Language Processing (NLP) systems on clinical data, including tasks for extracting complex relations from clinical narratives and electronic health records. The relevant entity types include medications/drugs and signs and symptoms of the patients. Medication-related entities include dosage, frequency, and type of medication. Relations identify whether a medication relieves or causes a symptom, whether the symptoms are new or part of the patients medical or family history, and so on. On the other hand, the BioNLP Shared Tasks [26–29] have focused on the extraction of fine-grained biological events and relations, such as gene regulation and protein–protein interactions, from the scientific literature. While these types of tasks do not generally use standardized vocabularies to ground their entities, their focus is on fostering the development of algorithms that prioritize semantic understanding of the text. Finally, we need to mention the drug–drug interaction corpus [30], which targeted the extraction of drug–drug interaction information from medical texts, an essential task for pharmacovigilance and patient safety; however, the task was localized at the sentence level, and most algorithms therefore depended on whether the interacting entities were mentioned in the same sentence.
In addition to the above-mentioned resources produced as benchmark datasets for the various community challenges and focus on relation annotation between biomedical entities, we were also inspired and encouraged by the significant efforts in the biocuration community to meticulously annotate and provide valuable resources for recognition of entities in the biomedical domain, such as the Colorado Richly Annotated Full-Text corpus [31] which covers many entity types, namely, disease [32], chemical [33, 34], gene [35], variant [36], cell line [37], and others. These corpora have been invaluable in the development of the current highly accurate tools in named entity recognition [38].
All the above-mentioned initiatives highlight the interdisciplinary effort towards leveraging NLP to extract meaningful relationships from vast and complex biomedical texts, aiding in the acceleration of knowledge discovery and healthcare advancements. However, the publicly available corpora have certain constraints: most of them do not annotate semantic relationships between entities of multiple types, or at the document level, often the annotated entity is not linked to a database concept identifier or a standardized vocabulary, and often, the annotation of relations does not distinguish relationships between novel findings and background or previously known associations. The development of relation extraction tools that are capable of addressing these challenges requires manually annotated corpora that contain (i) multiple biomedical entity types, (ii) relationships between them, (iii) sufficient examples in the document text, and (iv) enough articles for system training and an accurate evaluation of their performance. The BioRED-BC8 corpus, which supported the BioRED track at BioCreative VIII, is a rich corpus, manually annotated to respond to these challenges.
As shown in the overview of the BioRED track at BioCreative VIII, the challenge [39] aimed to foster development of multientity extraction systems capable of distinguishing between different relation types, as well as understanding from the context whether the extracted relationship is novel or background information. In fact, the BioRED initiative started in 2021. The original BioRED corpus [40] was a collection of 600 PubMed abstracts pooled from previously annotated corpora [32, 35, 41–43] rich in gene, disease, variant, and chemical entity mentions and enriched with the complete manual annotation for all chemical, gene, disease, species, gene variants and species entities mentioned in the abstracts. The biomedical entities were then equipped with their normalized identifiers to the corresponding vocabularies/ontologies: for diseases and chemicals—MeSH, for genes and proteins—NCBI Gene, for species—NCBI Taxonomy, for cell lines—Cellosaurus [37], and for gene/protein variants—dbSNP. Furthermore, the manual annotation included the pairwise relationships between those entities, the semantic type of relationship (i.e. positive correlation, negative correlation, binding, conversion, drug interaction, comparison, cotreatment, and association), and the novelty factor, namely, identifying whether the relationship constitutes a novel finding in the article or whether it is part of the background knowledge. This collection was used in the LitCoin challenge in 2022 (https://ncats.nih.gov/funding/challenges/winners/litcoin-nlp/details) and later on for the development of the BioREx [44] biomedical relation extraction tool.
During the BioCreative VIII BioRED track, an additional test dataset of 400 PubMed articles was selected to complement the original articles. These articles were selected to be rich in all the above-mentioned biomedical entities and pairwise relationships, with a similar distribution and composition to the training dataset. Additionally, it was designed to simulate a real-life application, and to reflect the current research trends in biomedical literature, the articles were selected from the literature recently published between 2021 and 2022. Each article was doubly annotated in a three-round annotation process, where annotator discrepancies were discussed after each round until the full consensus was reached. Figure 2 shows an example article in the BioRED-BC8 corpus and a snapshot of the entity and relationship annotations for that article. The BioRED-BC8 corpus, therefore, is an invaluable resource consisting of 1000 PubMed articles, with no restricting conditions on their sharing and distribution, fully available for the development of algorithms targeting relationship extraction in the biomedical domain; we expect it to be a significant contribution to this research field.
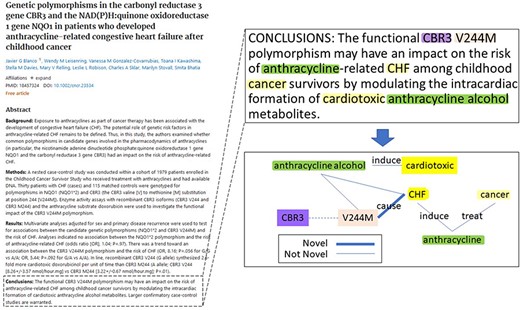
An example of article annotation data in the BioRED-BC8 corpus.
Taken together, this work makes the following significant contributions:
First, we present a new high-quality, manually annotated corpus on relations for the biomedical literature. The BioRED-BC8 corpus contains 1000 PubMed articles (title and abstract), and it differs from previous corpora because it was selected to be rich in biomedical entities and representative of relations between genes, diseases, chemicals, and variants. Also, these articles were published in many different journals to represent a large space of language variation. These characteristics make this corpus invaluable for the advancement and improvement of text-mining tools.
Second, we note that there is an end-to-end system that benefits from the entity identification tools that we have previously developed and included in the publicly available portal for all PubMed literature, PubTator. PubTator now includes the upgraded BioREx tool, which has been improved with training on this larger corpus and demonstrates better accuracy on relation extraction between biomedical entities. This tool is used daily to process the PubMed articles, and its results are publicly available at https://www.ncbi.nlm.nih.gov/research/pubtator3/.
Finally, we are interested in providing a practical use for our research. Our new resource, BioRED-BC8, is available at the CodaLab location https://codalab.lisn.upsaclay.fr/competitions/16381, where it continues to receive interest, and we believe that it will continue to be used as a benchmark to evaluate better systems in biomedical relation extraction.
This manuscript is organized as follows: in the Methods section, we describe our corpus development, annotation process, annotation guidelines, annotation tool, and the benchmark relation extraction method. In the Results section, we detail the corpus characteristics, and we compare these with the previous corpus and detail the new advantages. Finally, our evaluation shows that the new corpus can safely be used to test systems in real-life application simulations, where the testing data reflect the current trends in the biomedical literature. As shown, the results are robust, demonstrating that the model can generalize well to new unseen data.
Materials and Methods
Document selection procedure
The biomedical relationships corpus of the BioRED BioCreative VIII track had the following targets:
be representative of biomedical literature publications that contain biomedical mentions and biomedical relationships;
target articles that have no restrictions in sharing and distribution; and
be instrumental in training biomedical relation extraction algorithms to produce high-quality results even in new, never-before-seen articles;
To select the articles most suitable for algorithm testing, we focused on recently published articles. In a real-life application setting, the most useful algorithms are those that prove to be the most valuable for the incoming flux of never-before-seen published literature. As we experienced with the Coronavirus disease 2019 pandemic, correctly identifying what is discussed in the articles, and grouping those articles by the relevant topics, is most crucial, especially in the race to find an effective cure and a timely vaccine.
The 400 PubMed articles that constituted the BioRED track test set were selected to be as similar as possible to the BioRED original set of 600 articles, to be complementary, balanced, and a suitable test set, which can also serve as a standalone corpus (BioRED-BC8). The selection criteria included the following: maximization of journal coverage to ensure variety, a similar distribution of all entity mentions and identifiers per article, a similar distribution of all biomedical relationships per article, and similar language models.
Annotation guidelines
The complete BioRED corpus annotation guidelines are publicly available [40]. Here, we give a quick summary.
The guidelines specify which biomedical entities are considered, which entity pairs are considered, which semantic type categories are used for annotation, how to assign the entity pairs to the corresponding semantic category, and how to decide on a relationship novelty factor based on the article context. It also gives specific examples for each case.
The BioRED annotation team focused on six entity types which are commonly found in the biomedical literature: genes, chemicals, diseases, variants, species, and cell lines. Annotation for each of these entities was based on the works of previous research that developed well-known corpora focused on each of these entities, mainly, (i) genes, using the annotation guidelines of the NLM Gene corpus [35] and, accordingly, including mentions of genes, proteins, mRNA, and other gene products, normalized to the NCBI Gene database identifiers; (ii) chemicals, using the annotation guidelines of the NLM Chem corpus [34], which included mentions of chemical and drugs, normalized to the MeSH thesaurus identifiers; (iii) diseases, based on the annotation guidelines of the NCBI disease corpus [32] and accordingly covering mentions of diseases, symptoms, and some disease-related phenotypes, normalized to MeSH thesaurus identifiers, (iv) variants, using the annotation guidelines of the tmVar corpus [36] and including genomic/protein variants such as substitutions, deletions, insertions, and others, normalized to the dbSNP database identifiers; (v) species, inspired by the annotation guidelines of previous gene mention corpora which need to correspond closely to the species and organism mentions in the article, normalized to the NCBI Taxonomy database; and finally, (vi) cell lines, using the Cellosaurus as the normalizing vocabulary, and the annotation guidelines were based on the corpus annotation for the BioCreative VI Bio-ID track [45]. These standardized vocabularies and ontologies, if needed, can be easily mapped to other terminologies via the Unified Medical Language System (UMLS) [46].
In general, both the full mention and the abbreviated terms for each entity are annotated with the appropriate identifier for all occurrences in the title and abstract of an article. Composite mentions are annotated with a set of concepts in one-to-one correspondence with the mentions, i.e. the mention of the two genes in the text ‘MMP-1, -2’ is annotated as ‘4312, 4313’. For each annotation, the most specific term was preferred, so annotations to the high-level mentions of diseases such as ‘genetic disorder and cis-acting disease’ were discouraged and not included. In the case of conflicting or overlapping entity types, the most specific term was preferred, i.e. in ‘Adrenoleukodystrophy gene’, we annotated the gene, whereas in the ‘Adrenoleukodystrophy patient’, we annotated the disease.
Given the relations, due to the limited time and resources and the curation complexities with the multiple entity types, the BioRED project annotations only included binary relations and focus only on the most observed entity pairs as occurring in the biomedical literature, as well as the most common semantic types. These relation pairs are as follows: disease–gene, chemical–gene, disease–variant, gene–gene, chemical–disease, chemical–chemical, chemical–variant, and variant–variant. The semantic types are as follows: positive correlation, negative correlation, binding, conversion, drug interaction, comparison, cotreatment, and association. Other relation annotation projects have annotated more specific categories such as expression and activity; however, due to the curation complexity challenges in annotating multiple entities and multiple entity pair relations, as well as time constraints, we focused on a reduced set of semantic categories that are, to a certain extent, applicable over different pairs of entities:
A positive correlation includes the following: (a) an upregulation event between two genes; (b) an increased activity, exhibition, or response event between a chemical and a gene; (c) a response (or sensitivity) event between a chemical and a variant; (d) a cause between a gene and a disease; (e) when the presence of a chemical induces a disease, and (vi) when the presence of a chemical increases the activity of another chemical.
A negative correlation includes the following: (a) a downregulation event between two genes, (b) when a chemical suppresses the expression of a gene or the expression of a gene increases the resistance to a certain chemical, (c) when a resistance event is observed between a chemical and a variant, (d) when a negative correlation is observed between a gene and a disease, (e) when a chemical treats a disease, or (f) when a chemical inhibits the activity of another chemical.
A binding relation is observed between two genes or a chemical and a gene.
The relations of type conversion, drug interaction, comparison, and cotreatment are observed between two chemicals or drugs.
An association relation has a broader meaning and coverage. While every possible pair of our listed entity types is expected to be in an association relation, specific associated entity pairs could also have specific meanings, such as (a) an association relation between two genes may mean a regulation, modification, or other association events; (b) an association between a disease and a variant means that the inheritance of the genetic disease is confirmed by the variant; (c) when an association between a variant and a disease is confirmed, the corresponding gene is also associated with the disease; (d) a variant is associated with a disease if lower prevalence of the disease or lower disease susceptibility is observed in the presence of the variant; and (e) any modification event, e.g. phosphorylation, dephosphorylation, acetylation, deacetylation, and other modifications. Finally, we annotate the relation type as association for any relation between two entities that cannot be categorized as a positive/negative correlation or does not have a clear enough description to be categorized as another category.
In addition, different from previous corpora, the BioRED project aimed to annotate all the bioentity relations pertinent to a given article (title and abstract) that signified important semantic content for that article. This meant that relations between two given entities were not constrained to both entities co-occurring within the same sentence. Figure 3 illustrates such a relation from an article in the BioRED-BC8 corpus.
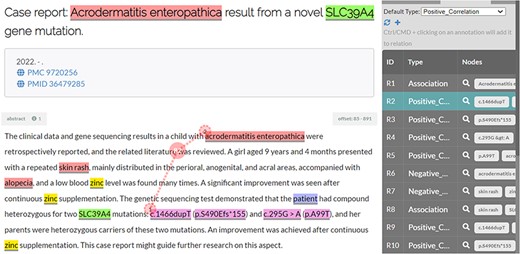
Illustration of a disease–variant relation (positive correlation) where the two entities do not co-occur in the same sentence.
Creating high-quality guidelines that fit the annotation task required a multistep iterative process, starting from an initial draft that was revised until clear and refined guidelines were obtained. The guidelines were prepared by professional expert annotators with degrees in chemistry, biochemistry, biological sciences, and molecular biology and with ample experience in annotating the PubMed literature.
Annotation procedure for the BioRED-BC8 resource
The BioRED-BC8 test articles are doubly annotated by eight NLM experts in two annotation phases: Phase 1, focusing on the annotation of entities, and Phase 2, focusing on the annotation of relationships. Each phase consisted of three annotation rounds. All manual annotations were performed using the TeamTat annotation tool [47].
For Phase 1, all articles were preannotated with entity mentions and their corresponding identifiers using PubTator Central [48], and for Phase 2, all articles were preannotated with BioREx, a state-of-the-art relation extraction method. For each phase, in the first round of annotations, each annotator worked independently, reviewed, edited the annotations as shown in the annotation tool, added new annotations that were missed, or deleted erroneous annotations, as needed. In the second round of annotations, each annotator reviewed all assigned articles that included their own annotations and their partner’s annotations of Round 1. In this step, they could make unbiased decisions on the annotated differences between the two partners since they were unaware of their partner’s identity. In the third annotation round, the annotator identities were revealed to their partners, and they discussed each article until they reached 100% consensus.
In addition, to further facilitate the work of the annotators, during the relationship annotation phase, a specific visualization tool was built to help visualize the differences in the annotation of the relationships as suggested by each annotator. This visualization showed the pair of entities in a relationship, the relationship type and the novelty factor for each annotated relationship, and included visual cues to identify the agreement between annotators and any remaining differences.
Corpus document format
While annotations can be represented in various formats, the BioRED-BC8 dataset is available in both BioC [49] (Extensible Markup Language and JavaScript Object Notation formats) and PubTator formats. These formats were chosen because they provided the following advantages: they support annotations representing both mention span (location) and entity identifier; articles in the PubMed Central (PMC) text-mining subset [50] are available in the BioC format; the TeamTat tool uses the BioC format; the text-mining tools we used for preannotation support these formats; PubTator Central Application Programming Interface (API) allows retrieval of any PubMed/PMC Open Access articles with precomputed annotations in any of these formats; and finally, these formats are simple and easy to modify, allowing additional analysis tools to be applied rapidly as needed.
BioCreative VIII BioRED task
The BioRED track at BioCreative VIII workshop consisted of two distinct subtasks:
Relation extraction (Subtask 1): Given a document (journal title and abstract) annotated with all mentions of entities, the participants are asked to identify the pairs of entities in a relationship and the semantic type of the relationship and its novelty factor, i.e. whether the relation constitutes a new result, conclusion, or observation of the current study or whether it is prior knowledge.
End-to-end system (Subtask 2): Given only the text of the document (journal title and abstract), the participants are asked to build an end-to-end system and identify the asserted relationships, their semantic relation type, and the novelty factor.
The BioRED training dataset was made available in April 2023, and the registered teams submitted their systems’ results in September 2023. The workshop was held in November 2023, co-located with the AMIA 2023 Annual Symposium. Fourteen teams worldwide submitted a total of 56 system output results for Subtask 1, and nine teams worldwide submitted 38 system output results for Subtask 2. These results are summarized in the BioRED track overview paper [39].
Benchmarking method
We used the method described in [40] to evaluate the corpus. This method was published 1 year prior to the BioCreative VIII challenge and as such made for an adequate benchmark method. The relation extraction BioRED model was built on a PubMedBERT implementation. PubMedBERT treats both subtasks as multiclass classification tasks. Given a pair of entities represented by their normalized IDs and the article text (title and abstract), PubMedBERT predicts a classification using the [CLS] tag. The BioRED model, in this context, produces two outputs: the relation type label and the novelty prediction label. The difference in the application of this model for Subtask 2 is that instead of the human annotations of the entity labels and their identifiers, we use the entity predictions as extracted from the PubTator APIs https://www.ncbi.nlm.nih.gov/research/pubtator3/.
Results and discussion
Corpus characteristics
The BioRED-BC8 track relationship extraction corpus is rich in bioentity mention and relationship annotations and currently the largest corpus, compatible with previously annotated corpora. The BioRED-BC8 training dataset of 600 PubMed articles contains 20 419 manual biomedical mention annotations; corresponding to 6502 manually annotated pairwise relationships. The BioRED-BC8 test dataset of 400 PubMed articles contains 15 400 manual biomedical mention annotations, corresponding to 6034 manually annotated pairwise relationships.
Figures 4 and 5 illustrate the entity and the relationship composition of the BioRED training and test datasets and demonstrate that the resources are (i) compatible—to foster reuse and acknowledge and build on previous efforts of experts and (ii) complementary—to expand on previous knowledge and cover new areas of training data.
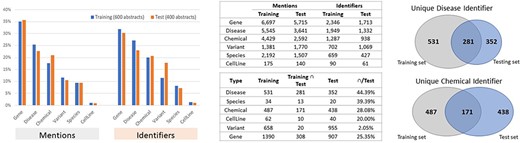
Entity composition in the BioRED-BC8 training and test datasets. The bar graphs illustrate the compatibility: the corpus composition for each relation type is relatively similar for both the training and testing datasets. The Venn diagrams illustrate that the training and testing datasets are complimentary, in that while a proportion of entities are present in both sets of articles, additional new data are available for each type.
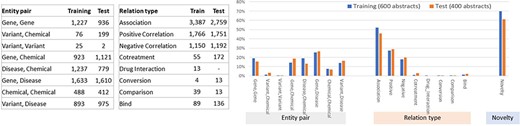
Relationship composition in the BioRED-BC8 training and test datasets.
Corpus technical validation
Table 1 shows the results of our benchmark method on Subtask 1. Our benchmark is based on our previously published method with the original BioRED corpus. The first implementation was trained on 500 articles as the training data and tested on 100 BioRED articles designated as the test data. This was the original LitCoin challenge setting. The second implementation used the complete 600 articles as the training dataset and the BioCreative-BC8 BioRED test data as the test dataset (400 articles). The new test data consist only of recently published articles and contain previously unseen entities, so differences in performances are expected. As expected, the performance is robust, while leaving room for further improvement.
Subtask 1 results when applying our PubMedBERT-based benchmark method on the BioRED (2021 version) test set and the BioRED-BC8 test dataset. ALL: Entity pair + relation type+ novelty
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| BioRED test | 76.13 | 75.41 | 75.77 | 58.51 | 57.95 | 58.23 | 64.58 | 63.97 | 64.28 | 48.52 | 48.07 | 48.29 |
| BioRED-BC8 test | 72.03 | 76.71 | 74.29 | 50.34 | 53.61 | 51.93 | 54.54 | 58.08 | 56.25 | 37.71 | 40.16 | 38.90 |
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| BioRED test | 76.13 | 75.41 | 75.77 | 58.51 | 57.95 | 58.23 | 64.58 | 63.97 | 64.28 | 48.52 | 48.07 | 48.29 |
| BioRED-BC8 test | 72.03 | 76.71 | 74.29 | 50.34 | 53.61 | 51.93 | 54.54 | 58.08 | 56.25 | 37.71 | 40.16 | 38.90 |
Subtask 1 results when applying our PubMedBERT-based benchmark method on the BioRED (2021 version) test set and the BioRED-BC8 test dataset. ALL: Entity pair + relation type+ novelty
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| BioRED test | 76.13 | 75.41 | 75.77 | 58.51 | 57.95 | 58.23 | 64.58 | 63.97 | 64.28 | 48.52 | 48.07 | 48.29 |
| BioRED-BC8 test | 72.03 | 76.71 | 74.29 | 50.34 | 53.61 | 51.93 | 54.54 | 58.08 | 56.25 | 37.71 | 40.16 | 38.90 |
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| BioRED test | 76.13 | 75.41 | 75.77 | 58.51 | 57.95 | 58.23 | 64.58 | 63.97 | 64.28 | 48.52 | 48.07 | 48.29 |
| BioRED-BC8 test | 72.03 | 76.71 | 74.29 | 50.34 | 53.61 | 51.93 | 54.54 | 58.08 | 56.25 | 37.71 | 40.16 | 38.90 |
Table 2 shows the results of Subtask 2. In this task, we needed to first identify the entities mentioned in the titles and abstracts of the articles in the test dataset. To perform this step, we utilized both the current and the newest versions of PubTator, specifically PubTator 2.0 and PubTator 3.0 [38]. As displayed, better entity recognition predictions result in better relation extraction performance. As expected, the BioRED-BC8 test dataset is an adequate test dataset that complements the BioRED training dataset.
Subtask 2 results when applying our PubMedBERT-based benchmark method on the BioRED (2021 version) test set, and the BioRED-BC8 test dataset
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| PubTator 2.0—BioRED test | 55.06 | 33.71 | 41.81 | 43.26 | 26.48 | 32.85 | 43.26 | 26.48 | 32.85 | 32.58 | 19.95 | 24.75 |
| PubTator 2.0—BioRED-BC8 | 45.28 | 37.12 | 40.80 | 32.10 | 26.32 | 28.92 | 33.96 | 27.84 | 30.60 | 24.12 | 19.77 | 21.73 |
| PubTator 3.0—BioRED test | 57.40 | 41.36 | 48.08 | 44.99 | 32.42 | 37.68 | 46.78 | 33.71 | 39.18 | 35.92 | 25.88 | 30.08 |
| PubTator 3.0—BioRED-BC8 | 55.49 | 47.98 | 51.46 | 39.67 | 34.30 | 36.79 | 41.69 | 36.05 | 38.66 | 29.76 | 25.73 | 27.60 |
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| PubTator 2.0—BioRED test | 55.06 | 33.71 | 41.81 | 43.26 | 26.48 | 32.85 | 43.26 | 26.48 | 32.85 | 32.58 | 19.95 | 24.75 |
| PubTator 2.0—BioRED-BC8 | 45.28 | 37.12 | 40.80 | 32.10 | 26.32 | 28.92 | 33.96 | 27.84 | 30.60 | 24.12 | 19.77 | 21.73 |
| PubTator 3.0—BioRED test | 57.40 | 41.36 | 48.08 | 44.99 | 32.42 | 37.68 | 46.78 | 33.71 | 39.18 | 35.92 | 25.88 | 30.08 |
| PubTator 3.0—BioRED-BC8 | 55.49 | 47.98 | 51.46 | 39.67 | 34.30 | 36.79 | 41.69 | 36.05 | 38.66 | 29.76 | 25.73 | 27.60 |
Because in Subtask 2, we are not given the entity annotations, we use the PubTator 2.0 and PubTator 3.0 tool outputs as our entity predictions.
Subtask 2 results when applying our PubMedBERT-based benchmark method on the BioRED (2021 version) test set, and the BioRED-BC8 test dataset
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| PubTator 2.0—BioRED test | 55.06 | 33.71 | 41.81 | 43.26 | 26.48 | 32.85 | 43.26 | 26.48 | 32.85 | 32.58 | 19.95 | 24.75 |
| PubTator 2.0—BioRED-BC8 | 45.28 | 37.12 | 40.80 | 32.10 | 26.32 | 28.92 | 33.96 | 27.84 | 30.60 | 24.12 | 19.77 | 21.73 |
| PubTator 3.0—BioRED test | 57.40 | 41.36 | 48.08 | 44.99 | 32.42 | 37.68 | 46.78 | 33.71 | 39.18 | 35.92 | 25.88 | 30.08 |
| PubTator 3.0—BioRED-BC8 | 55.49 | 47.98 | 51.46 | 39.67 | 34.30 | 36.79 | 41.69 | 36.05 | 38.66 | 29.76 | 25.73 | 27.60 |
| . | Entity pair . | Entity pair + relation type . | Entity pair + novelty . | ALL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | P . | R . | F . | P . | R . | F . | P . | R . | F . | P . | R . | F . |
| PubTator 2.0—BioRED test | 55.06 | 33.71 | 41.81 | 43.26 | 26.48 | 32.85 | 43.26 | 26.48 | 32.85 | 32.58 | 19.95 | 24.75 |
| PubTator 2.0—BioRED-BC8 | 45.28 | 37.12 | 40.80 | 32.10 | 26.32 | 28.92 | 33.96 | 27.84 | 30.60 | 24.12 | 19.77 | 21.73 |
| PubTator 3.0—BioRED test | 57.40 | 41.36 | 48.08 | 44.99 | 32.42 | 37.68 | 46.78 | 33.71 | 39.18 | 35.92 | 25.88 | 30.08 |
| PubTator 3.0—BioRED-BC8 | 55.49 | 47.98 | 51.46 | 39.67 | 34.30 | 36.79 | 41.69 | 36.05 | 38.66 | 29.76 | 25.73 | 27.60 |
Because in Subtask 2, we are not given the entity annotations, we use the PubTator 2.0 and PubTator 3.0 tool outputs as our entity predictions.
Discussion
The corpus for the BioRED track at BioCreative VIII is a high-quality dataset and consists of (i) the training dataset, previously published as BioRED, that contains 600 PubMed abstracts and (ii) the test dataset that contains 400 recently published PubMed abstracts, specifically annotated for the BioCreative VIII challenge. The BioRED-BC8 corpus thus contains 1000 PubMed articles, fully annotated for diseases, genes, chemicals, species, gene variants, and cell lines. All entities are also annotated with their corresponding database identifiers: NCBI Gene for genes and proteins, NCBI Taxonomy for species, dbSNP for gene/protein variants, MeSH for diseases and chemicals, and Cellosaurus for cell lines. Furthermore, all articles are fully annotated for pairwise relationships between disease–gene, chemical–gene, disease–variant, gene–gene, chemical–disease, chemical–chemical, chemical–variant, and variant–variant and their semantic categories: positive correlation, negative correlation, binding, conversion, drug interaction, comparison, cotreatment, and association. Finally, each relationship is categorized for novelty, distinguishing whether that relationship is a significant finding for that article or whether it describes previously known facts or background knowledge. The BioRED-BC8 corpus is a gold-standard corpus for biomedical relation extraction in biomedical articles.
To identify any potential pitfalls and help summarize key points for future method improvements, we selected all articles with false-positive and false-negative predictions and manually reviewed one-tenth of these articles. The challenges of the relation extraction task can be broadly grouped into the below categories. Note that an error may be affected by more than one challenge:
Implicit, insufficient, or ambiguous statements: Approximately 55.6% of the total errors were due to implicit, insufficient, or ambiguous statements describing the relationship in the article text. These types of sentences are often very long, which causes the method to have difficulty in identifying the key verb indicating the relation. For example, in the article with PubMed Identifier (PMID):31829566, ‘Following injection, only afucosylated “N-glycan” structures were passed through enzyme zones that contained alpha2-3 sialidase, followed by beta1-3,4 galactosidase, which cleaved any terminal alpha2-3-linked sialic acid and underlying galactose yielding a terminal “N-acetyl glucosamine”’. The relation between ‘N-glycan’ and ‘N-acetyl glucosamine’ is explained through a series of chemical reactions. The statement is too long to understand easily, causing our method to miss the relation. As another example, in PMID: 22 476 228, ‘rs17833172 and rs3775067 SNPs of the ADD1 gene and the rs4963516 SNP of the GNB3 gene were significantly associated with the BP response to CPT, even after adjusting for multiple testing’, the variants of the ADD1 and GNB3 genes are related, implying a relation between the ADD1 and GNB3 genes. However, since no direct relation is explicitly stated in the text, our method missed the relation between the two genes.
Relation across multiple sentences: We found that ∼8.5% of the total errors consisted of entities mentioned far apart in the text and in different sentences. Since the BioRED task corpus is curated at the document level, many entity pairs are not mentioned within the same sentences, increasing the difficulty of the relation extraction task. For example, in PMID: 22476228, ‘Genetic factors influence blood pressure (BP) response to the cold pressor test (CPT), which is a phenotype related to “hypertension” risk… Haplotype analysis indicated that the CCGC haplotype of ADD1 constructed by rs1263359, rs3775067, rs4961, and rs4963 was significantly associated with the BP response to CPT’, the ADD1 gene and its variants are related to hypertension due to their genetic influence on blood pressure (BP) and the cold pressor test (CPT). The relationship between the ADD1 variants and hypertension is conveyed across multiple sentences, thereby increasing the difficulty of the relation extraction task.
Conflicting statements in different sentences of the same document: This type of error accounted for ∼6.5% of the total errors of our benchmark method. For example, in PMID: 31416966, ‘Blocking the interaction between “AHR” and “kynurenine” with CH223191 reduced the proliferation of colon cancer cells’, and ‘We found that only “kynurenine” and no other tryptophan metabolite promotes the nuclear translocation of the transcription factor “aryl hydrocarbon receptor (AHR)”’, the first sentence mentions an interaction between AHR and kynurenine but does not specify the type of correlation. The following sentence clearly states that kynurenine promotes the nuclear translocation of AHR, representing a positive correlation. Such situations may cause inconsistent judgements by the deep learning model, leading to incorrect relation-type errors.
Arguable statements: This type accounts for ∼15.2% of the errors. The entity pairs are mostly in a grey area and also created arguments during human curation. For example, in PMID: 27623776, ‘Like “carvedilol, nebivolol” reduced the opening of single RyR2 channels and suppressed spontaneous Ca2+ waves in intact hearts and “catecholaminergic polymorphic ventricular tachycardia (CPVT)” in the mice harboring a RyR2 mutation (R4496C)’, the sentence describes that the carvedilol, nebivolol suppressed spontaneous Ca2+ waves in the mice with R4496C. The model missed the relations between ‘carvedilol/nebivolol’ and R4496C. However, the statement in the text is not clear. In another example, PMID: 31298765 states, ‘SSBP1 mutations in dominant optic atrophy with variable retinal degeneration’, the relation between SSBP1 and ‘optic atrophy’ is incorrectly recognized, since the discussion in the paper primarily focuses on the relationship between SSBP1 and ‘Autosomal Dominant Optic Atrophy’, rather than its broader category, ‘optic atrophy’. Such arguable cases can lead to false positives or false negatives.
n-Ary relations: Another challenging category (6.4%) involves relations that do not simply involve only two entities. According to the current annotation guidelines, the n-ary relations are annotated as binary relations between all corresponding pairs; however, such annotations may not be easily identifiable. For example, in PMID :27374774, ‘Complexome profiling recently identified TMEM126B as a component of the mitochondrial complex I assembly complex alongside proteins ACAD9, ECSIT, NDUFAF1, and TIMMDC1.’ The curators annotated every pair of genes named in this complex as an association relation, which were missed by our method. Additionally, difficulty is observed in capturing complex relations involving an interaction between two entities which is conditional on specific circumstances or when another entity is involved in the interaction. For example, consider PMID: 27784654, ‘Furthermore, CALCB increases ERK1/2 phosphorylation in a time-dependent manner in RUASM, and the protective effect of CALCB on TNF-alpha-induced inhibition of CALCRL/RAMP1 associations was significantly blocked in presence of ERK inhibitor (PD98059).’ In this article, CALCB positively correlates with CALCRL and RAMP1; however, this involves a negative correlation between TNF-alpha and the CLACRL/RAMP1 complex.
Others: A total of 11.3% of errors were due to a combination of various reasons which cannot be grouped in any of the above categories. We believe that additional training data for the machine to learn the background knowledge behind biomedical entity relations could help with many such cases. For example, in PMID: 33278357, ‘/’ in ‘Mechanistically, TNF-alpha and IFN-gamma co-treatment activated the JAK/STAT1/IRF1 axis’ represents co-work in the same pathway. However, the model does not understand the meaning of ‘/’ since it was not adequately learned during the training dataset.
Gold-standard data are crucial for the development of robust models, and we demonstrated that in the BioRED-BC8 corpus, both training and test datasets have matching entity composition and distributions and matching relationship composition and distributions. The training and test datasets introduce new, previously unseen elements, and they complement each other to create a larger, richer gold-standard dataset for semantic relation extraction in biomedical journals. We tested the new corpus with our best performing relation extraction tool, and for Subtask 2, we paired it with both PubTator 2.0– and PubTator 3.0–predicted entities.
Conclusion
The BioRED-BC8 relationship corpus provides the following contributions: (i) high-quality manual annotation of six types of biomedical entities; (ii) all entity mentions are normalized to standardized vocabularies and ontologies, which, via UMLS, can be easily mapped to other terminologies, if needed, (iii) document-level semantic annotation of pairwise relationships; and (iv) novel relations based on the article context. Annotations were performed by the expert literature biocurators at the NLM. The annotation guidelines are compatible with previously annotated corpora; therefore, these corpora can be used as additional data. We believe that this freely available resource will be invaluable for advancing text-mining techniques for biomedical relation extraction tasks in biomedical text.
Conflict of interest
None declared.
Funding
This research was supported by the NIH Intramural Research Program, NLM. It was also supported by the National Natural Science Foundation of China (No. 62302076 to L.L.).
Data availability
The BioRED-BC8 corpus is available at https://codalab.lisn.upsaclay.fr/competitions/16381. Using the CodaLab setup allows for all users to use the same evaluation scripts as the participants in the BioCreative VIII BioRED track challenge. The leaderboard allows a true evaluation between any submitted system that uses the same dataset, thus providing a true benchmarking setting and relieving individual researchers or research groups from the burden of downloading and managing different systems to compare with their own results. For the BioCreative VIII challenge, the BioRED track CodaLab website was used 383 times, and since the BioCreative VIII challenge, in November 2023, the leaderboard has received more than 250 submissions from all over the world.



