-
PDF
- Split View
-
Views
-
Cite
Cite
Cao Hengchun, Guo Hui, Yang Weifei, Li Guiting, Ju Ming, Duan Yinghui, Tian Qiuzhen, Ma Qin, Feng Xiaoxu, Zhang Zhanyou, Zhang Haiyang, Miao Hongmei, SesamumGDB: a comprehensive platform for Sesamum genetics and genomics analysis, Database, Volume 2024, 2024, baae105, https://doi.org/10.1093/database/baae105
Close - Share Icon Share
Abstract
Sesame (Sesamum indicum L., 2n = 26) is a crucial oilseed crop cultivated worldwide. The ancient evolutionary position of the Sesamum genus highlights its value for genomics and molecular genetics research among the angiosperms of other genera. However, Sesamum is considered a small orphan genus with only a few genomic databases for cultivated sesame to date. The urgent need to construct comprehensive, curated genome databases that include genus-specific gene resources for both cultivated and wild Sesamum species is being recognized. In response, we developed Sesamum Genomics Database (SesamumGDB), a user-friendly genomic database that integrates extensive genomic resources from two cultivated sesame varieties (S. indicum) and seven wild Sesamum species, covering all three chromosome groups (2n = 26, 32, and 64). This database showcases a total of 352 471 genes, including 6026 related to lipid metabolism and 17 625 transcription factors within Sesamum. Equipped with an array of bioinformatics tools such as BLAST (basic local alignment search tool) and JBrowse (the Javascript browser), SesamumGDB facilitates data downloading, screening, visualization, and analysis. As the first centralized Sesamum genome database, SesamumGDB offers extensive insights into the genomics and genetics of sesame, potentially enhancing the molecular breeding of sesame and other oilseed crops in the future.
Database URL: http://www.sgbdb.com/sgdb/
Introduction
Sesame (Sesamum indicum L., 2n = 26) is an ancient and crucial oilseed crop, notably known for its high oil content and quality, and is prominent worldwide [1, 2]. Sesame, which originated in Africa and is now widely cultivated in the tropical and subtropical regions of Asia, Africa, and South America, has a history of cultivation dating back to ∼3000–50 bc in the Harappa Valley of the Indian subcontinent [3]. The genus Sesamum, classified within the Pedaliaceae family, is notable for its diversity, comprising over 30 wild species in addition to the single cultivated species S. indicum L [4, 5]. Compared with the cultivated sesame, some wild species, such as Sesamum calycinum, Sesamum latifolium, and Sesamum radiatum, exhibit higher resistance and tolerance to biotic and abiotic stresses, making them valuable genetic resources for advancing modern molecular breeding in sesame [6].
With the rapid advancement of sequencing technologies, substantial progresses in sesame genomics and molecular breeding have been achieved over the past decade. Notably, the completion of the Sesame Genome Project (SGP) in 2020 provided essential genome data for Sesamum genomics research [7, 8]. Pan-genome analysis of chromosome-scaled genomes from the cultivated sesame and six wild Sesamum species provided clear evidence of genome evolution and domestication. For the Sesamum genus, four chromosome groups (i.e. groups A, B, C, and D) were named and differentiated for the first time. Both whole-genome duplication and whole-genome triplication events were identified in the assembled seven Sesamum species. A typical allotetraploidization event occurred in S. radiatum (2n = 4x = 64), significantly contributing to the divergence of species within the Sesamum genus [3]. Genome evolution analysis revealed losses and expansions in numerous gene families that regulate key agronomic traits and crucial processes, such as plant type, inflorescence meristem development, fatty acid biosynthesis and metabolism, and resistance to Fusarium wilt disease in sesame [3]. To date, multiple de novo assembled genome maps of cultivated sesame (four varieties) and seven wild species have been published [9–12].
To enhance the accessibility of sesame genome data, various raw sequencing data, and genomics information, several sesame genome databases have been established. These databases can be categorized into three groups on the basis of their primary resources. The first category includes online genome databases, such as Sinbase and the SGP database (http://sesamum.org/), which focus on de novo assembled genome data and were constructed in 2015 and 2013, respectively [13]. The second category, reported between 2015 and 2021, comprises databases dedicated to sesame functional genomics, including SesameFG, SiGeDiD, and SesameHapMap [14–16]. The third category includes molecular marker databases, such as GinMicrosatDb and SisatBase, which were also published during this period [17, 18]. However, all these genome databases cover only the cultivated sesame, and most are not continuously accessible by users worldwide.
In this study, we integrated the latest genomic resources from the SGP with additional public datasets, which include two cultivated sesame varieties (Yuzhi11 and Zhongzhi13) and seven wild Sesamum species. Our objective was to develop a novel and user-friendly genome database for Sesamum genus, named Sesamum Genomics Database (SesamumGDB). This database includes a suite of practical bioinformatics online tools and provides a comprehensive genomic resource that focuses on specific gene families, including lipid biosynthesis and metabolism, and transcription factors (TFs). We identified and systematically cataloged 6026 genes related to lipid metabolism and 17 625 TF genes across 58 types in 8 Sesamum species. This comprehensive platform is poised to make significant contributions to sesame genomics research and enhance molecular breeding efforts in the future.
Materials and methods
Data sources
The database encompasses nine high-quality de novo genomic datasets, representing two cultivated sesame varieties, Yuzhi 11 and Zhongzhi 13 (S. indicum, 2n = 2x = 26), and seven wild species, namely S. alatum, S. angustifolium, S. latifolium, S. calycinum, and S. angolense (all 2n = 2x = 32), along with the tetraploid species S. radiatum and S. schinzianum (both 2n = 4x = 64). A comprehensive overview is provided in Table 1, which details all genomic data that are made publicly accessible.
Catalog of genome information of two cultivated sesame varieties and seven wild relatives in SesamumGDB
| Species . | Variety name . | Type . | Karyotype . | Genome size (Mb) . | N50 (Mb) . | Gene number . | NCBI accession . | Reference . |
|---|---|---|---|---|---|---|---|---|
| S. indicum | Yuzhi11 | Cultivated | 2n = 26 | 346.8 | 22.9 | 31 462 | GCA_003268515.1 | [6] |
| S. indicum | Zhongzhi13 | Cultivated | 2n = 26 | 270.3 | 17.3 | 35 410 | GCF_000512975.1 | [7] |
| S. alatum | 3651 | Wild | 2n = 26 | 528.0 | 40.5 | 25 722 | GCA_034509735.1 | [3] |
| S. angustifolium | G01 | Wild | 2n = 32 | 300.7 | 11.6 | 32 646 | JACGWK000000000 | |
| S. latifolium | Ken1 | Wild | 2n = 32 | 369.0 | 22.2 | 42 114 | JACGWN000000000 | |
| S. calycinum | Ken8 | Wild | 2n = 32 | 313.0 | 14.3 | 31 417 | JACGWM000000000 | |
| S. angolense | K16 | Wild | 2n = 32 | 300.8 | 24.1 | 31 091 | GCA_034509725.1 | |
| S. radiatum | G02 | Wild | 2n = 64 | 668.4 | 19.5 | 68 397 | JACGWJ000000000 | |
| S. schinzianum | Gangguo | Wild | 2n = 64 | 704.5 | 19.7 | 54 212 | GCA_027475655.1 | [10] |
| Species . | Variety name . | Type . | Karyotype . | Genome size (Mb) . | N50 (Mb) . | Gene number . | NCBI accession . | Reference . |
|---|---|---|---|---|---|---|---|---|
| S. indicum | Yuzhi11 | Cultivated | 2n = 26 | 346.8 | 22.9 | 31 462 | GCA_003268515.1 | [6] |
| S. indicum | Zhongzhi13 | Cultivated | 2n = 26 | 270.3 | 17.3 | 35 410 | GCF_000512975.1 | [7] |
| S. alatum | 3651 | Wild | 2n = 26 | 528.0 | 40.5 | 25 722 | GCA_034509735.1 | [3] |
| S. angustifolium | G01 | Wild | 2n = 32 | 300.7 | 11.6 | 32 646 | JACGWK000000000 | |
| S. latifolium | Ken1 | Wild | 2n = 32 | 369.0 | 22.2 | 42 114 | JACGWN000000000 | |
| S. calycinum | Ken8 | Wild | 2n = 32 | 313.0 | 14.3 | 31 417 | JACGWM000000000 | |
| S. angolense | K16 | Wild | 2n = 32 | 300.8 | 24.1 | 31 091 | GCA_034509725.1 | |
| S. radiatum | G02 | Wild | 2n = 64 | 668.4 | 19.5 | 68 397 | JACGWJ000000000 | |
| S. schinzianum | Gangguo | Wild | 2n = 64 | 704.5 | 19.7 | 54 212 | GCA_027475655.1 | [10] |
Catalog of genome information of two cultivated sesame varieties and seven wild relatives in SesamumGDB
| Species . | Variety name . | Type . | Karyotype . | Genome size (Mb) . | N50 (Mb) . | Gene number . | NCBI accession . | Reference . |
|---|---|---|---|---|---|---|---|---|
| S. indicum | Yuzhi11 | Cultivated | 2n = 26 | 346.8 | 22.9 | 31 462 | GCA_003268515.1 | [6] |
| S. indicum | Zhongzhi13 | Cultivated | 2n = 26 | 270.3 | 17.3 | 35 410 | GCF_000512975.1 | [7] |
| S. alatum | 3651 | Wild | 2n = 26 | 528.0 | 40.5 | 25 722 | GCA_034509735.1 | [3] |
| S. angustifolium | G01 | Wild | 2n = 32 | 300.7 | 11.6 | 32 646 | JACGWK000000000 | |
| S. latifolium | Ken1 | Wild | 2n = 32 | 369.0 | 22.2 | 42 114 | JACGWN000000000 | |
| S. calycinum | Ken8 | Wild | 2n = 32 | 313.0 | 14.3 | 31 417 | JACGWM000000000 | |
| S. angolense | K16 | Wild | 2n = 32 | 300.8 | 24.1 | 31 091 | GCA_034509725.1 | |
| S. radiatum | G02 | Wild | 2n = 64 | 668.4 | 19.5 | 68 397 | JACGWJ000000000 | |
| S. schinzianum | Gangguo | Wild | 2n = 64 | 704.5 | 19.7 | 54 212 | GCA_027475655.1 | [10] |
| Species . | Variety name . | Type . | Karyotype . | Genome size (Mb) . | N50 (Mb) . | Gene number . | NCBI accession . | Reference . |
|---|---|---|---|---|---|---|---|---|
| S. indicum | Yuzhi11 | Cultivated | 2n = 26 | 346.8 | 22.9 | 31 462 | GCA_003268515.1 | [6] |
| S. indicum | Zhongzhi13 | Cultivated | 2n = 26 | 270.3 | 17.3 | 35 410 | GCF_000512975.1 | [7] |
| S. alatum | 3651 | Wild | 2n = 26 | 528.0 | 40.5 | 25 722 | GCA_034509735.1 | [3] |
| S. angustifolium | G01 | Wild | 2n = 32 | 300.7 | 11.6 | 32 646 | JACGWK000000000 | |
| S. latifolium | Ken1 | Wild | 2n = 32 | 369.0 | 22.2 | 42 114 | JACGWN000000000 | |
| S. calycinum | Ken8 | Wild | 2n = 32 | 313.0 | 14.3 | 31 417 | JACGWM000000000 | |
| S. angolense | K16 | Wild | 2n = 32 | 300.8 | 24.1 | 31 091 | GCA_034509725.1 | |
| S. radiatum | G02 | Wild | 2n = 64 | 668.4 | 19.5 | 68 397 | JACGWJ000000000 | |
| S. schinzianum | Gangguo | Wild | 2n = 64 | 704.5 | 19.7 | 54 212 | GCA_027475655.1 | [10] |
Genome reannotation
To ensure the consistency and reliability of gene annotation and prediction across the nine Sesamum genomes, we conducted a comprehensive reannotation. Tandem repeats in each genome were identified using Tandem Repeats Finder (v4.09) with default settings [19]. Transposable elements were detected using LTR_finder (v1.07), LTR_retriever (V2.9.0), and RepeatModeler (v2.0.3) [20–22]. Whole-genome repeat sequences were then masked using RepeatMasker (v4.1.1) [23, 24]. Gene structure prediction was combined with ab initio-, homology-, and RNA-seq-based methods. Braker2 and tblastn were employed for ab initio- and homology-based gene predictions, respectively [25].
We predicted homologous proteins between the assembled Sesamum genomes and four model plants closely related to sesame, including Arabidopsis thaliana, Solanum lycopersicum, Vitis vinifera L., and Mimulus guttatus (Supplementary Table S1). This was performed using tblastn, with an e-value of 1e-5. Hisat2 (v2.1.0), Stringtie (v1.3.4), and PASA (v2.3.3) were used for RNA-seq-based gene prediction [26, 27]. Furthermore, EVidenceModeler was used to integrate the results of gene predictions and obtain a consensus gene set [28]. Throughout the gene prediction process, the Non-Redundant Protein Sequence Database, Kyoto Encyclopedia of Genes and Genomes (KEGG), SwissProt, and Gene Ontology (GO) databases were comprehensively used for gene function annotation [29–32]. Noncoding RNAs, including rRNAs, small RNA, cis-regulatory elements, and tRNA, were identified using a combination of Infernal (v1.1.2), tRNAscan-SE (v2.0.10), and RNAmmer programs and the Rfam database [33, 34].
Gene family identification
To accurately identify and functionally annotate gene families within the Sesamum genomes, we employed two methodologies: emapper and pfam_scan. The primary protein sequences from the eight species were annotated using emapper (v2.1.7) and pfam_scan (v1.6), both set to their default parameters [35, 36].
TF identification
The web tools in the Plant Transcription Factor Database (PlantTFDB) were used to identify TFs in the nine genomes [37]. We conducted the TF predictions using the protein sequences derived from these genomes on the Transcription Factor Prediction page, adhering to the default settings to maintain consistency. After completing the prediction process, we systematically organized and enumerated the identified TFs according to their nomenclature, facilitating a clear and structured classification of the results.
Identification of genes related to lipid metabolism
The online tool Mercator4 (v6.0) was used to identify genes related to lipid biosynthesis and metabolism in the Sesamum genomes [38]. We uploaded the protein sequences through the FASTA (Fast All Sequences in A) validation process and utilized the default settings on the protein annotation interface to ensure a standardized analysis. Once the analysis was complete, we engaged in a thorough review of the results, selectively identifying genes that fell under the “Lipid metabolism” category. Subsequently, these genes were methodically categorized based on their involvement in the specific subprocesses of lipid metabolism, allowing for a systematic and detailed classification of the identified genes related to lipid pathways.
Integration of bioinformatics tools with the SesamumGDB
We integrated the SequenceServer with the SesamumGDB to deliver a robust BLAST (basic local alignment search tool) service and provide a user-friendly experience. As an advanced front-end, the SesamumGDB could be used to perform sequence alignment and search [39]. Furthermore, the latest version of JBrowse (the Javascript browser) 2 was used for constructing SesamumGDB and comprehensively visualizing all accessible genomes [40].
SesamumGDB implementation
The SesamumGDB was developed using the classic LAMP (Linux + Apache + MySQL (My Structured Query Language)+ PHP) stack within a RedHat system on Centos 7.3 environment. This database provides user-friendly web pages for data searching and browsing. The web interface is built with HTML5 and PHP (v5.4) and runs on an Apache web server. The backend is supported by MariaDB (v5.5.68), which houses several interrelated relational databases, including those for lipid metabolism-related genes. The database interfacing and the common gateway interface are programmed in Perl. SesamumGDB is hosted on a World Wide Web server, offering internet access via a web client. The browsers recommended for accessing the database are Google Chrome and Internet Explorer 10.0+ (or higher).
Results
Utilizing a diverse dataset encompassing nine genomes and employing an advanced search platform, we developed SesamumGDB, a pioneering genome database dedicated to the Sesamum genus (Fig. 1a). SesamumGDB provides extensive genomic information on individual genes, gene families, TFs, and genes related to lipid biosynthesis and metabolism. Moreover, this database features a robust search catalog that facilitates data downloads, searches, and browsing, including homolog BLAST, GO term, and KEGG pathway screening. In addition, the platform supports dataset visualization and navigation, presenting all data and search results in a user-friendly interface (Fig. 1b). The detailed architecture and functionalities of SesamumGDB are presented in the subsequent sections.
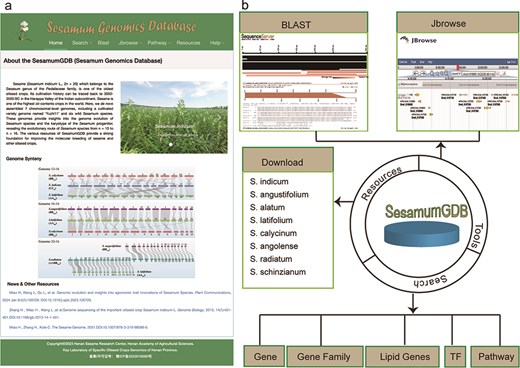
Overview of SesamumGDB, inculding the homepage screenshot (a) and the foundational functions and useful tools (b).
Details of SesamumGDB
SesamumGDB includes 352 471 genes derived from 9 genomes, involving those of 2 cultivated sesame varieties (Yuzhi 11 and Zhongzhi 13) and 7 wild Sesamum species (Table 1). To ensure the reliability of genome information, all nine genomes were reannotated, incorporating data from the GO and KEGG databases, providing a richer context for gene functions and pathways. The database contained a vast array of 317 385 gene families (Supplementary Table S4). Given that sesame is a crucial oilseed crop, special attention was paid to gene families related to energy storage and regulation. Using the Mecrator software for protein annotation, 6026 genes associated with lipid biosynthesis and metabolism were identified (Supplementary Table S3). This comprehensive categorization serves as a valuable resource for further research, which can facilitate the exploration of evolutionary connections and functional commonalities among these gene families in Sesamum.
The data underwent detailed refinement, enabling systematic screening and cataloging of TF genes across the nine genomes (Fig. 2, Supplementary Table S2). A total of 17 625 TF entries were classified into 58 categories. This curated collection highlights the key regulatory components that play a crucial role in the complex regulation of gene expression in Sesamum.
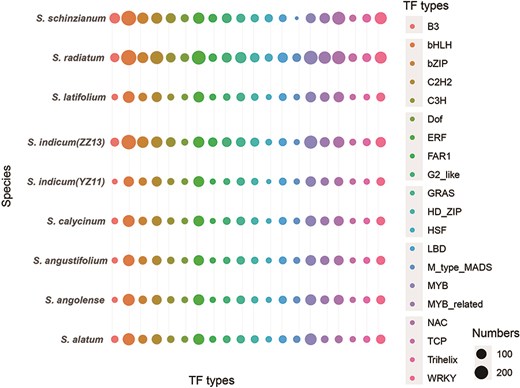
Searching and browsing genes and genomic features
To enhance user access to Sesamum genome data and highlight the advantages of the SesamumGDB database, we developed four search terms and integrated them into the “Search” menu. Users can retrieve information on Sesamum-specific genes, lipid-related genes, TF genes, and gene families by using the Gene Retrieve module (Fig. 3). In the “Gene Retrieve” module, users can explore genes by species classification or by entering specific gene IDs. The primary output page provides an exhaustive overview of each gene, including details such as gene ID, species, variety, genomic location, strand, and comprehensive annotations from Pfam, GO, and KEGG databases. Moreover, the database displays the description of genes and their corresponding nucleotide and protein sequences (Fig. 3a).
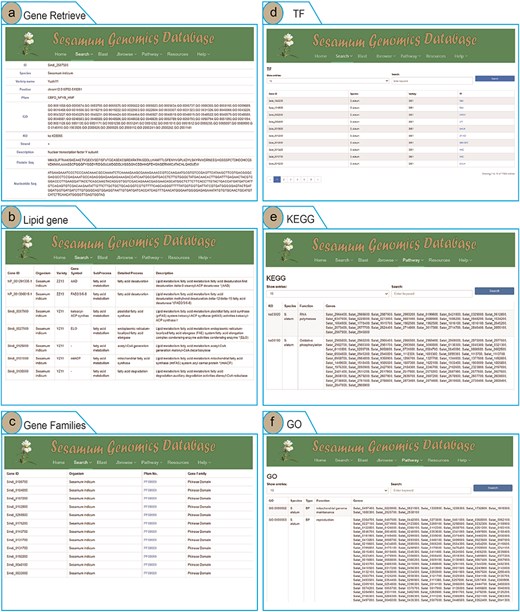
Genomic feature and exploration examples of SesamumGDB, including gene retrieve by species name and gene ID (a), targeted search for lipid metabolism genes (b), gene family discovery by species and Pfam classification (c), TFs in Sesamum genomes (d), KEGG pathways browsing (e), and GO terms annotations survey (f).
The search functions in the “Lipid Genes” and “Gene Families” modules are similar, allowing users to select a species and enter specific subtypes of lipid metabolism pathways or gene families into the provided text field. Upon submitting a query, the system generates a detailed report of the search results (Fig. 3b and c).
In the “Transcription Factor” module, users can access all TFs by searching via gene IDs, species, variety, or TF names (Fig. 3d). The interface also allows users to customize the number of entries displayed per page. Additionally, by clicking on the TF names, users can directly hyperlink to the PlantTFDB website, where they can retrieve detailed information regarding the specific TF protein [37].
Homolog blast
We integrated the SequenceServer software (version 2.0.0) into SesamumGDB to provide a user-friendly interface for conducting homolog BLAST analyses and visualizing the results (Fig. 4a). Users can either paste their query sequence(s) directly or drag and drop a file containing the sequence(s) in the FASTA format into the designated input field. They can then select from a range selection of databases specifically curated for homology search, each constructed using the coding DNA sequences (CDS) and protein sequences from the nine genomes housed within SesamumGDB. The BLAST analysis was streamlined with other programs (BLASTN, BLASTP, BLASTX, tBLASTN, and tBLASTX) and automatically configured to match the nature of the submitted query sequence and the selected database. The platform offers default parameter settings; however, users can customize these settings through an “Advanced parameters” box, allowing them to adjust the e-value threshold, scoring matrix, and output formatting to meet their specific research needs. The alignment results are then elegantly presented using various visualization techniques. These visual representations, which are informative and available for downloaded in both scalable vector graphics and Portable Network Graphics (PNG) formats, are of high-quality and can be used directly for presentations and further analysis.
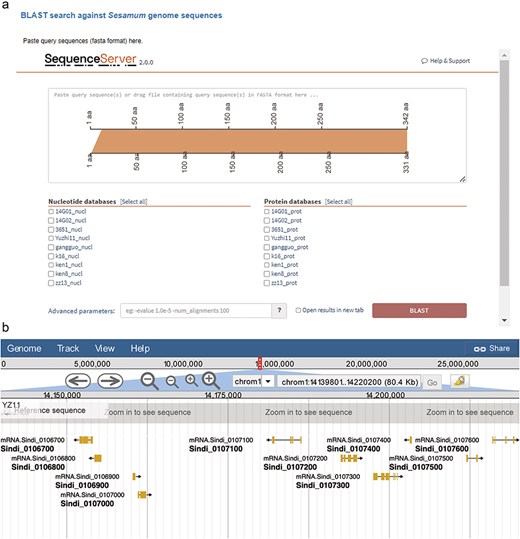
Integrated bioinformatic platforms in SesamumGDB, featuring SequenceServer for gene-specific searches (a) and JBrowse for detailed Sesamum genome annotation review (b).
JBrowse
Apart from searching by gene IDs or genomic locations, SesamumGDB enables the advanced exploration of nine high-quality Sesamum genomes through the integration of JBrowse 2. Due to its ability to rapidly visualize and navigate large-scale genomic datasets, JBrowse 2 enhances user interaction with each genome featured in the SesamumGDB (Fig. 4b). Various implemented tracks facilitate the browsing of key genomic features, including genome sequences, CDSs, protein-coding gene models, and exon regions. Users can conveniently isolate a specific chromosome for in-depth examination of the genomic features. The interactive nature of JBrowse 2 facilitates the straightforward selection of elements within any given track. Once an element is selected, an information panel instantly appears to the right of the genome browser interface, presenting the essential details such as gene name, position, length, and sequences. This user-centric design ensures a streamlined experience catering to the diverse needs of the research community.
GO terms and KEGG pathways
In the SesamumGDB, the “Pathway” menu includes GO and KEGG modules, both of which are crucial for elucidating the functions of protein-coding genes. All genes across the nine genomes were annotated using the eggNOG database (v 5.0.2), allowing for the construction of a comprehensive set of GO terms and KEGG pathways for Sesamum genes [41]. The database presents a total of 100 719 GO terms for 153 690 genes and 1296 KEGG pathways for 76 171 genes (Supplementary Table S5).
The “KEGG” module in SesamumGDB offers a straightforward search mechanism, where users can input keywords such as gene ID, species name, KEGG Orthology (KO) number, or functions derived from biological experiments and bioinformatics analysis. These inputs facilitate the rapid retrieval of KEGG-related data. All results are displayed in a tabular format (Fig. 3e). Additionally, by clicking on a KO number, users can access a direct hyperlink to the KEGG Pathway Database (https://www.kegg.jp/kegg/pathway.html) [31], where a detailed pathway map for the specified KO number is presented.
The “GO’”module in SesamumGDB parallels the functionality of the “KEGG” module, allowing users to retrieve GO terms and directly access more detailed information via hyperlinks to the AmiGO2 database for a specific GO term [42]. This streamlined approach ensures easy accessibility to gene function annotation information and the broader biological context covered by these databases (Fig. 3f).
Data download
To ensure comprehensive access to the genomic resources, SesamumGDB includes a “Resource” page where all processed data related to the nine sesame genomes are readily available for download. These data include a wide range of genomic sequences, CDS, complementary DNA, protein sequences, and detailed annotations. To enhance the downloading experience and to ensure swift data retrieval, all datasets were systematically organized and stored in the compressed zip file format. This approach not only ensures a fast download but also allows researchers to efficiently manage and access the extensive genomic information presented in the SesamumGDB.
A case study of the application of SesamumGDB
To illustrate the main functions of the database, we present a case study of the NAC (NAM/ATAF/CUC) gene in the SesamumGDB. The NAC gene Sindi_2309500 was identified as crucial in modulating the upregulation of the oil content in sesame [3]. In the “Gene Retrieve” section, the profile of Sindi_2309500 is organized into 11 categories (Fig. 5a). This gene is located on chromosome 10 of the sesame genome (var. Yuzhi 11), spanning from positions 15 393 884 bp to 15 395 322 bp and orientated on the negative strand. Pfam analysis has identified a “No apical meristem” (NAM) domain within this gene, and it is annotated as a NAC domain-containing protein [43, 44]. In addition, the profile includes detailed information regarding the gene’s nucleotide sequences and its corresponding amino acid composition.
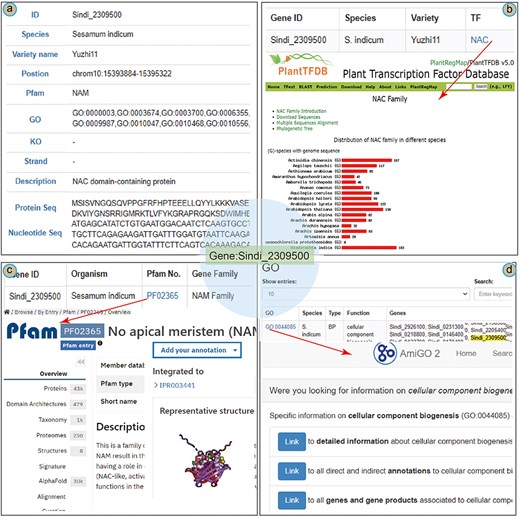
A case study of Sindi_2309500 gene in SesamumGDB, including gene profiles (a), transcription factor discovery (b), gene family exploration (c), and GO term search with database cross-referencing (d).
As a TF, detailed information on Sindi_2309500 is accessible through the “Transcription Factor” module (Fig. 5b). The exact match results include a hyperlink to the TF term “NAC,” providing an extensive overview of the NAC family based on PlantTFDB. These results elucidate the characteristics and distribution of the NAC encoded by Sindi_2309500 across the Sesamum species. Furthermore, entering “NAC” into the search function yields a complete list of all 1001 NAC genes across the nine Sesamum genome.
Regarding Sindi_2309500, the gene search results highlighted the presence of a NAM domain (PF02365) in its protein sequence as a notable feature (Fig. 5a). This NAM domain could be further explored by navigating to the “Gene Families” module. Here, “NAM family” was entered as the search keyword, and “Sesamum indicum” was selected to specify the species. This targeted search approach yielded a succinct list of all 72 genes containing the NAM domain within the S. indicum genome. Additionally, clicking on the direct link to the InterPro database with the specific Pfam identifier [45], provided more detailed insights into the NAM domain’s functional and structural attributes (Fig. 5c).
In addition to basic search functionalities, the “GO” module in the SesamumGDB allows users to enter the specific gene identifiers to obtain detailed gene function information. For instance, upon entering the gene ID Sindi_2309500, we could retrieve a comprehensive list of associated GO terms, identifying a total of 41 distinct GO terms for S. indicum (var. Yuzhi11). By selecting an individual GO term, such as GO:0044085, users are directed to the AmiGO2 website where detailed information on the GO:0044085 term is presented (Fig. 5d).
The KEGG module of the SesamumGDB presents detailed pathway maps related to specific genes in a tabular format. For example, because Sindi_2309500 lacks KEGG annotation, we randomly selected another gene, Sindi_0007900, for exploration within this module. When Sindi_0007900 was entered into the KEGG module, a set of five results were displayed in a tabular format. By clicking on a KO number in the table, such as ko00061, we obtained a detailed pathway map in KEGG database (Supplementary Fig. S1). In this case, the pathway map corresponded to the fatty acid biosynthesis pathway, illustrating the significance of SesamumGDB in the exploration of both function and related metabolic pathways associated with a gene. Furthermore, to ensure the effective utilization of SesamumGDB, we have provided a comprehensive tutorial accessible under the “Help” menu.
Discussion and conclusion
Sesame, a crucial oilseed crop, occupies a unique position in the genome evolution of eudicot plants [3, 46]. In this study, we developed SesamumGDB, the first genome database specifically curated for the genus Sesamum. This database compiles genomic information for the single cultivated species and seven wild relatives. Moreover, this database lays the foundation for future pan-genome analyses and functional genomics and genetics research in sesame, especially considering the rich agronomic traits that have emerged through domestication and selective breeding processes [47]. Thus, the extensive genomic sources of Sesamum are expected to help resolve genetic bottlenecks associated with sesame domestication [48, 49].
As the first database for Sesamum, the SesamumGDB offers an extensive collection of genomic data from nine genomes across eight species, covering all three chromosome groups found in Sesamum (2n = 26, 32, and 64). Unlike existing sesame genome databases that primarily focus on the cultivated sesame species, the SesamumGDB provides a wealth of genomic information for the entire Sesamum genus. This includes comprehensive data on gene families, with a particular emphasis on crucial genes related to lipid biosynthesis and metabolism, as well as TFs (Fig. 1b).
Our commitment to updating the genome datasets has significantly facilitated gene discovery and genomic analysis for sesame. We anticipate an influx of more comprehensive sesame genome data in the near future. SesamumGDB, as an essential tool, is well-positioned to evolve and integrate these new genomic resources for sesame. We are dedicated to continuously updating the database and enhancing the robustness of genome information in the SesamumGDB, establishing it as a cornerstone for global sesame research.
Supplementary data
Supplementary data is available at Database online.
Conflict of interest
None declared.
Funding
This work was financially supported by Key Research Project of the Shennong Laboratory (SN01-2022-04), the earmarked fund for China Agricultural Research System of MOF and MARA (CARS-14), Henan Province Specific Professor Position Program (SPPP2023), Zhongyuan Scientist Workshop Construction Fund (092101211100), the Innovation Scientist and Technician Troop Construction Project of Henan Province (ISTTCPHP2016), and the Science and Technology Innovation Team Project of Henan Academy of Agricultural Sciences (2023TD04), Science and Technology Foundation for The Excellent Youth Scholars of Henan Academy of Agricultural Sciences (2022YQ14), and Scientific and Technological Project of Henan Province (242102110254).



