-
PDF
- Split View
-
Views
-
Cite
Cite
Romain Guyot, Perla Hamon, Emmanuel Couturon, Nathalie Raharimalala, Jean-Jacques Rakotomalala, Sreenath Lakkanna, Sylvie Sabatier, Antoine Affouard, Pierre Bonnet, WCSdb: a database of wild Coffea species, Database, Volume 2020, 2020, baaa069, https://doi.org/10.1093/database/baaa069
Close - Share Icon Share
Abstract
Coffee is a beverage enjoyed by millions of people worldwide and an important commodity for millions of people. Beside the two cultivated species (Coffea arabica and Coffea canephora), the 139 wild coffee species/taxa belonging to the Coffea genus are largely unknown to coffee scientists and breeders although these species may be crucial for future coffee crop development to face climate changes. Here we present the Wild Coffee Species database (WCSdb) hosted by Pl@ntNet platform (http://publish.plantnet-project.org/project/wildcofdb_en), providing information for 141 coffee species/taxa, for which 84 contain a photo gallery and 82 contain sequencing data (genotyping-by-sequencing, chloroplast or whole genome sequences). The objective of this database is to better understand and characterize the species (identification, morphology, biochemical compounds, genetic diversity and sequence data) in order to better protect and promote them.
Introduction
Coffee is a beverage enjoyed by millions of people worldwide. It is also an important commodity for millions of small coffee farmers living in tropical countries. Two species are mainly cultivated Arabica (Coffea arabica) and Robusta (Coffea canephora). Coffee fields are deeply impacted by climate change and the emergence of diseases (1). Beside cultivated coffee trees, numerous wild coffee species are known to botanists, but largely ignored by agronomists and breeders although these species may be crucial for future coffee crop development to face climate changes.
Based on morphological data, the phylogenetically closest genus called Psilanthus (2) has been recently placed into Coffea (3). The genus Coffea (broad sense since (3)) includes woody plants belonging to the Rubiaceae family. It comprises 124 species and 17 additional taxa, with a natural distribution covering tropical Africa, Madagascar, Comoros, Mauritius and the Reunion Islands extending to Southern and Southeast Asia and Australasia. However, the two genera mainly differ by the flower morphology with short corolla tube and reproductive organs exerted for Coffea, long corolla tube and reproductive organs inserted for Psilanthus. They differ also by their natural distribution. The genus Psilanthus is present on the African continent, Asia (India, Sri Lanka, tropical and Southeast Asia) and Oceania (Northern Australia) but absent from the islands of the West Indian Ocean (Madagascar, Mascarenes and Comoros). The merging of these two genera introduced a possible source of name confusion as numerous works are focused only on Coffea sensu stricto. Another source of species name confusion is the presence of numerous past or modern synonymies. For example, it is generally accepted that Psilanthus minor is in fact P. sapinii or that Coffea vaughanii is C. myrtifolia.
Anyway, these 141 identified and classified species/taxa are currently considered for further phylogenetic and molecular analyses (4).
Fourteen years ago, Davis and co-workers (5) revealed that numerous wild coffee species are vulnerable (23 species), endangered (30 species) or seriously threatened (19 species). A recent reassessment (1) confirmed that 60% of them are now threatened with extinction, suggesting a bad prospect for wild coffee species all over the tropical world. Wild coffee species living collections were initiated in the years 1960 in Africa and Madagascar. Today only 55% of them are in such collections: at the research station of the Centre National de Recherche en Agronomie, Divo, Côte d’Ivoire (for African species), at the Centre de Ressources Biologiques (CRB) Bassin-Martin, Reunion island (http://florilege.arcad-project.org/fr/crb/coffea; for African, Comorian and Mascarene species; Supplemental data 1) and at the research station of the National Centre for Applied Research for Rural Development (FOFIFA) in Kianjavato, Madagascar (for Madagascan species; Supplemental data 2). The analysis of wild coffee species conserved in living collection revealed large morphological variation such as flower morphology, size and color of fruits, plant height and leaf morphologies, days to fruit maturation and growth habitats and adaptation. In addition to morphology, large variations were observed in terms of seed biochemical compounds involved in the quality of coffee such as caffeine (6), trigonelline, sucrose and mangiferin contents into others (7–9). However, this diversity is not comprehensively reported so far in any publication or any publicly available database.
The first Coffea genome has been published in 2014 and concerned Coffea canephora (10). Since this first release, several sequencing data have been published such as the genotyping-by-sequencing data to provide the first resolved phylogeny of the Coffea genus (11), the partial sequencing of 16 Coffea species (12) and the chloroplast reconstructions and nuclear SNP mining (13). Now the Genus is subjected to intensive genome sequencing (C. arabica, C. eugenioides and C. canephora (14); 82 wild coffee species, unpublished results; Supplemental data 3), allowing research of genes of agronomical interest and genome composition and evolution studies.
To intensify the protection and the conservation of wild coffee species and to promote their use in the search of agronomic genes of interest, more shared information is needed. So far, few databases are dedicated to wild coffee species. The Global Biodiversity Information Facility (GBIF, https://www.gbif.org/species/2895315) includes 8462 occurrences with images for 176 species. However, the reported samples include those collected in their natural area and also, those introduced by humans outside their natural distribution and those collected from ex situ living collections. The Reunion_Coffea database hosted in GBIF database was published by INRA Antilles-Guyane (15, https://www.gbif.org/dataset/510b8030-6293-4bd9-812d-c195a9915a74). This database provides the taxonomic distribution of occurrences (number of accessions per species), date of setting up in Reunion but does not include pictures or associated Supplementary data. Other databases are only focused on genomic data, such as the coffee Genome hub (http://coffee-genome.org; Robusta genome only), the TropGeneDB (http://tropgenedb.cirad.fr/tropgene/JSP/interface.jsp?module=COFFEE, mainly genetic markers) and the SOL Genomics Network https://sgn.cornell.edu/search/organisms).
In this study, we developed a wild coffee species database (WCSdb), hosted by Pl@ntNet (http://publish.plantnet-project.org/project/wildcofdb_en). The general objective of this database is to better understand the species (identification, morphology, biochemical compounds, genetic diversity and sequence data) in order to better protect and promote them. More specifically, the database presents: (i) each species held in collection on the sites of Bassin-Martin, Reunion Island and Kianjavato, Madagascar with a photo gallery of the tree morphology with a total of 597 images; (ii) different detailed information such as synonymy, natural distributions, habitats, architectural, morphological, phenological, biochemistry traits, genetic/genomic data, trait of interest retrieved from the literature and personal observations on living collection and (iii) a general geographical map of the species distribution.
Database construction and content
Data source
Pictures were mainly provided by Emmanuel Couturon for CRB (BassinMartin, La réunion) hosted plants (Supplemental data 1; list of species present at BassinMartin), by Eva N Raharimalala and Perla Hamon for Madagascan species (Supplemental data 2; list of species present at Kianjavato), and Lakkanna Sreenath for Psilanthus. bengalensis, Psilanthus. travancorensis and Psilanthus. wightianus. Links to molecular data were introduced such as genome size (Mbp 1C) from the plant DNA C-value database (https://cvalues.science.kew.org) for 57 species, genotyping-by-sequencing (GBS) data for 77 species (11, https://datadryad.org/resource/doi:10.5061/dryad.kk71t), the sequenced and assembled chloroplast genomes for 39 species (13; Guyeux et al. unpublished results) and whole genome deep pair-end Illumina sequencing for 82 species (R. Guyot and Perla Hamon, unpublished results, data available upon request). genotyping-by-sequencing (GBS) and sequencing data information are available on the web site in a separate table and in Supplemental data 3).
Web interface, usage
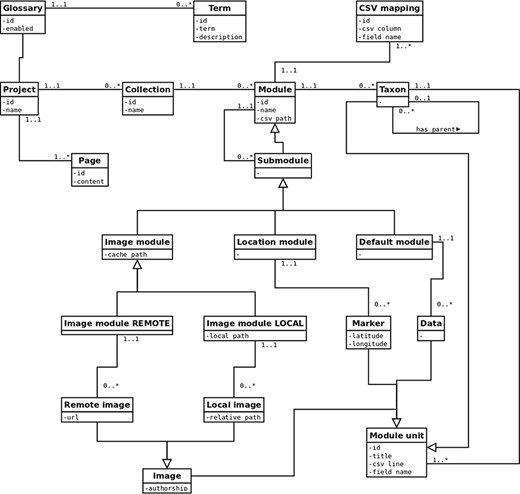
Pl@ntNet Publish is an IT platform dedicated to the dissemination of botanical data focused on taxa or specimen levels. It is based on Symfony (PHP) and MongoDB and allows its users to manage data publication spaces: descriptive texts, search forms, taxonomic data, visual data, geographic data, etc. (Figure 1). Data can be uploaded via comma-separated values (CSV) files and are then available through responsive Web pages dedicated to the portal created by users. Data exploration is also possible via a RESTful Web services (JSON). Pl@ntNet Publish was initially developed in 2014, with the support of Agropolis Fondation and has been adapted to various cases studies, such as Herbarium collections, Regional taxonomic checklists, information for Weed and Invasive species management, into others. Pl@ntNet Publish was deposited in 2015 at the Agency for the Protection of Programs (IDDN.FR.001.320007.OOO.R.C.2015.000.31235).
Schematic representation of the Pl@ntNet Publish UML class diagram.
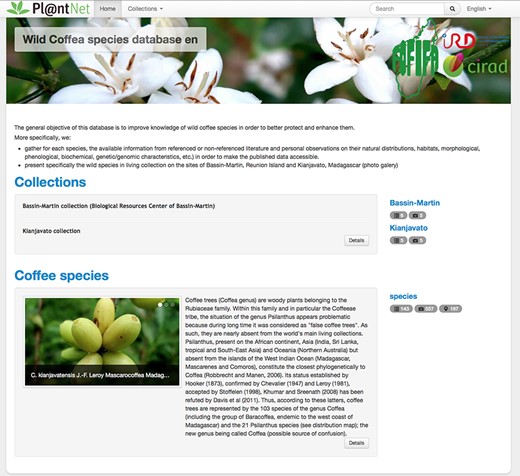
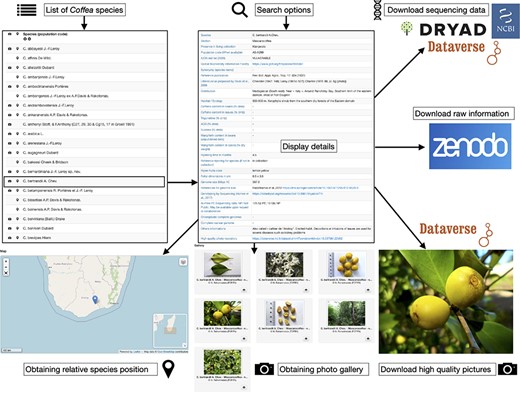
The main tab or species tab shows information for 124 accepted species and 17 taxa (Figure 2). Three species has duplicated entries, according to their distribution or variety: C. mauritiana for populations from Mauritius and Reunion islands, for Coffea liberica (variety liberica and variety dewevrei), and for Psilanthus bengalensis (variety bababudanii and variety bengalensis). In the species tab, we decide to keep the nomenclature Coffea and Psilanthus for the species to avoid any confusion. This database also shows species according to their botanical section or geographic origin: Eucoffea, Mascarocoffea, Mascarenes, Baracoffea (Madagascan species endemic to the western coast), Psilanthus from Africa and Psilanthus from Asia. The species table allows the user to select the species to examine. In total, for each species, 30 data fields were completed into a detailed table such as species name, section, living collection, synonymy and reference, distribution, habitat, caffeine content in bean and leaves, sucrose, mangiferin, ripening time and color of mature fruits, foliar dimension, genome sizes, sequencing data, and plastid genomes into others (Figure 3). The fields are described in the web site (section ‘How to use this site’). The detailed table contains also references and the links to access to the molecular data when available (GBS data, nuclear genome sequences, plastid genomes and raw sequencing data).
Home page of the wild coffee species database (http://publish.plantnet-project.org/project/wildcofdb_en).
In total, 201 localization entries were linked with coffee species, representative approximate area where the species were found when collected in the wild (mainly between years 1960 and 1980). With the detailed table, a photo gallery is available for 84 species. The photographs depict the morphology of beans, leaves, mature fruits, flowers, and the overall tree, allowing a better identification. In total, 551 photographs are available.
Conclusions and prospects
The WCS database represents the first comprehensive information about wild coffees species, largely unknown to coffee scientists and breeders. The information collected from the literature and from living collections at Kianjavato, Madagascar and Bassin-martin, La Réunion, may help researchers working in the preservation of coffee species, geneticists and breeders working with trait or genes of interest and improvement of cultivated species (tolerance to drought, resistances to diseases and pest, increase the quality of beans) or breeders motivated to re-cultivate forgotten species adapted to climate changes or adapted to specific habitats. More information will be integrated into the WCS database in the future, such as the availability of raw genomic sequences and assembled genome sequences, new characterized species, and new photo gallery.
Supplementary Data
Supplementary data are available at Database Online.
Funding
We thank the respective institutions IRD, FOFIFA, and CIRAD for support and funding.
References
Author notes
Citation details: Guyot,R., Hamon,P., Couturon,E., et al. WCSdb: a database of wild Coffea species Database (2020) Vol. 00: article ID baaa069; doi:10.1093/database/baaa069