-
PDF
- Split View
-
Views
-
Cite
Cite
Taner Z. Sen, Carson M. Andorf, Mary L. Schaeffer, Lisa C. Harper, Michael E. Sparks, Jon Duvick, Volker P. Brendel, Ethalinda Cannon, Darwin A. Campbell, Carolyn J. Lawrence, MaizeGDB becomes ‘sequence-centric’, Database, Volume 2009, 2009, bap020, https://doi.org/10.1093/database/bap020
Close - Share Icon Share
Abstract
MaizeGDB is the maize research community’s central repository for genetic and genomic information about the crop plant and research model Zea mays ssp. mays. The MaizeGDB team endeavors to meet research needs as they evolve based on researcher feedback and guidance. Recent work has focused on better integrating existing data with sequence information as it becomes available for the B73, Mo17 and Palomero Toluqueño genomes. Major endeavors along these lines include the implementation of a genome browser to graphically represent genome sequences; implementation of POPcorn, a portal ancillary to MaizeGDB that offers access to independent maize projects and will allow BLAST similarity searches of participating projects’ data sets from a single point; and a joint MaizeGDB/PlantGDB project to involve the maize community in genome annotation. In addition to summarizing recent achievements and future plans, this article also discusses specific examples of community involvement in setting priorities and design aspects of MaizeGDB, which should be of interest to other database and resource providers seeking to better engage their users. MaizeGDB is accessible online at http://www.maizegdb.org.
Database URL:http://www.maizegdb.org
Introduction
Maize is one of very few species that serve both as an important research model and as a crop from which diverse products and resources are generated [reviewed in (1, 2)]. This breadth of scope is recapitulated by the wide variety of informatics needs expressed by the community of maize biologists—not only are tools for handling genetic and genomic information needed, support for translational and applied research is also of great interest [reviewed in (3)].
To better understand the broad needs of the research community and prioritize development goals, a Working Group (http://www.maizegdb.org/working_group.php) made up of maize geneticists and computational biologists meets annually to discuss the MaizeGDB project’s status and to suggest how to further develop the MaizeGDB resource. In addition, the maize community periodically organizes meetings to gather information on key needs to move maize research forward. In March 2007, lab heads met at the Allerton Park and Conference Center in Monticello, IL, to discuss ‘The Future of Maize Genetics’ [meeting report available at http://www.maizegdb.org/AllertonReport.doc and (4)]. Guidance from the MaizeGDB Working Group and Allerton reports agree that two needs are of the utmost priority: improving access to the genome sequence of inbred line B73 (as well as other maize genome sequences as they become available) and creating tools to improve phenotype data collection, storage and analysis. With this in mind, sequence data and phenotypes constitute much of the current MaizeGDB Project Plan, a document that outlines work to be accomplished by MaizeGDB over a 5-year period (2009–14). In brief, the goals are as follows: MaizeGDB currently has a wide range of maize data including genetic maps, gene products, loci, alleles, phenotypes, stocks, sequences and markers. However, centralized access to currently ongoing maize projects that create sequence-indexed data (roughly 10–15 projects at any given time) is notably lacking. Reported here are some recent updates to MaizeGDB with emphasis on improving the handling and accessibility to sequence data, especially data generated by the Maize Genome Sequencing Project for B73 (6). Of particular note are (i) the new MaizeGDB Genome Browser (see ‘Genomic sequence data display and integration with genetic maps’ section), (ii) a new project ancillary to MaizeGDB called POPcorn, which currently serves as a portal to maize research projects with a centralized maize sequence similarity search resource coming soon and (iii) a recently launched project to involve the community of maize geneticists in genome annotation for B73 (outlined in ‘Current endeavors’ section).
to integrate new maize genetic and genomic data into the database by
expanding mutant and phenotype data and tools as well as structural and genetic map sets emphasizing the integration of the IBM genetic maps with the B73 genome sequence;
creating views that convey the substantial variation in maize genome structure;
integrating the next-generation genetic map being generated by the Maize Diversity Project (5) into a genomic view to enable its effective use by plant breeders;
providing access to gene models calculated by leading gene structure prediction groups through the MaizeGDB interface;
compiling and making accessible the annual Maize Newsletter at MaizeGDB
to provide community support services, such as lending help to the community of maize researchers with respect to developing and publicizing a set of guidelines for researchers to follow to ensure that their data can be made available through MaizeGDB; coordinating annual meetings; and conducting elections and surveys.
MaizeGDB’s standard operating procedures, machine architecture, accessibility and a description of how the databases are administered are described elsewhere (1, 2). Data made available via MaizeGDB are in the public domain.
Genomic sequence data display and integration with genetic maps
Genome browser
Based upon the 2006 MaizeGDB Working Group guidance (available at the bottom of http://www.maizegdb.org/working_group.php) and the Allerton meeting report (4), the MaizeGDB Team began development toward making MaizeGDB become more sequence-centric in early 2007. To this end, an initiative to implement a MaizeGDB Genome Browser was launched in early 2008 and completed in December 2008. The MaizeGDB Genome Browser enables MaizeGDB to become the long-term and centralized keeper of maize gene models (which ensures proper nomenclature) and serves as a way to compare various groups’ assemblies and annotations simultaneously.
A variety of genome browser applications were evaluated via a survey prepared on behalf of the Maize Genetics Executive Committee (accessible online at http://www.maizegdb.org/blanksurvey.html) to gauge cooperators' impressions of existing software and to find out what functionalities they would like to have in a maize genome browser. A summary of the survey results is available online at http://www.maizegdb.org/genome_browser_survey.php.
Based upon results of the survey, GBrowse (7) was selected for the following reasons: As the MOD for maize, MaizeGDB strives to consolidate data from any group that makes information available. For this reason, the MaizeGDB BAC-based Genome Browser (http://bac.maizegdb.org) has been populated with many groups’ data to create an integrated view of the maize genome within a single resource. Current contributors include the Maize Mapping Project [MMP (13)], the Maize Genome Sequencing Consortium [MGSC via their MaizeSequence.org resource (6)], PlantGDB (14), the UniformMu group (15), the Department of Energy’s Joint Genome Institute (http://www.phytozome.org/maize.php) and the Maize Assembled Gene Islands (MAGIs) resource (16). The MaizeGDB Genome Browser includes direct links to PlantGDB, MAGI and MaizeSequence.org from several ‘tracks’ allowing direct access to relevant data and tools available from those specialized resources.
The three most desired features reported were ease of use, visuals and speed. Cross-species comparison capabilities [where Ensembl (8) shines] was ranked fourth, and GBrowse has such capabilities available (SynBrowse (9), GBrowse syn [http://gmod.org/wiki/GBrowse_syn], CMap [reviewed in (10)], etc.).
Cooperators would like to see specific tool development to enhance their research (e.g. finding all the genes between two given markers). Flexibility of code and tool development is an intrinsic feature of GBrowse, which is a community-based open source project that allows development of new modules via its customizable plug-in architecture.
Those surveyed commented on various Model Organism Databases (MODs) and their genome browsers. Sites using GBrowse [e.g. TAIR and FlyBase; (11) and (12)] received far fewer negative comments than sites using other mainstream software platforms.
The B73 MMP and MGSC outcomes serve as the basis for the Genome Browser’s framework. The MGSC used a minimal tiling path of ∼19 000 mapped bacterial artificial chromosome (BAC) clones derived from the MMP for BAC-based genome sequencing. The focus has been to produce high-quality coverage of all identifiable gene-containing regions of the maize genome where only gene-containing regions are ordered, oriented and anchored to the physical and genetic maps of the maize genome (6).
For the MaizeGDB Genome Browser, genomic coordinates for elements that can be mapped to the B73 genome representation are provided by individual research groups, which creates a need for coordination in data release among the various contributors. To facilitate coordination, MaizeGDB, PlantGDB and MaizeSequence.org are currently working together to align features to the same GenBank (17) releases. Until this coordination is fully accomplished, genomic features displayed within the MaizeGDB Genome Browser may have been aligned by individual groups to different GenBank record releases, causing the elements on different tracks within a particular BAC to be out of order relative to each other. This is due to the fact that subsequences within each BAC can be reordered and reoriented as the records are updated in GenBank. Maize pseudomolecules have been computed (18) and will serve as the backbone that enables all groups to accomplish data coordination. A pseudomolecule-based Genome Browser at MaizeGDB is currently under preparation.
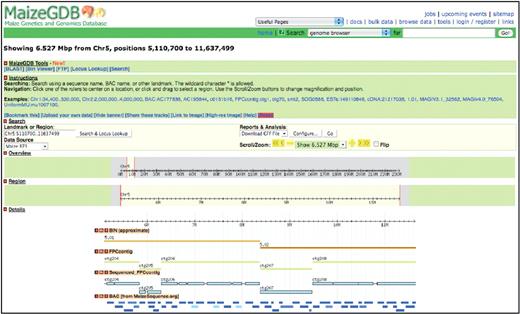
In addition to data provided by contributors, two large-scale tracks have been calculated and are included in the MaizeGDB Genome Browser: a ‘Sequenced FPC contig’ track that clearly specifies regions that are not yet sequenced and a ‘BIN’ track that relies upon association of genetic markers to BACs for estimating maize bin boundaries (Figure 1). In the ‘Sequenced FPC contig’ track, sequenced regions are shown as boxes and unsequenced regions are shown as lines. These representations are based upon mapping the BACs to the FPC contigs (13) and demonstrate places where the minimal tiling path does not fully cover the contig. For the ‘BIN’ track, the 90 genetic markers that delineate stretches of ∼10–20 cM along each chromosome (19) were mapped to the genome. These bin boundary markers are called the ‘core bin markers’. Because only a fraction of these core bin markers currently align with precision greater than a BAC, the displayed bin boundaries are only approximate. In some cases, there is no evidence (e.g. derived from sequence-based or hybridization-based methods) indicating bin boundary position. In such instances the corresponding bins are shown to extend between the flanking core bin markers. For example, although the core bin marker umc5a should delineate the boundary between bins 2.06 and 2.07 on chromosome 2, that marker sequence currently aligns only to a region on chromosome 7: the core bin marker for 2.07 is not known. This results in complete overlap of bins 2.06 and 2.07 within the browser.
The MaizeGDB Genome Browser showing chromosome 5 from nucleotide position 5 110 700 to 11 637 499. The ‘BIN’ track shows bins 5.01 and 5.02, and the ‘Sequenced FPC contig’ track clearly displays regions within the FPC contigs that are not currently sequenced.
Although the current view of the B73 genome is fluid and continues to change as more data become available, the addition of the genome browser to MaizeGDB enables researchers to use the emerging B73 sequence data in real time and allows them to visualize genomic elements, identify how these elements are positioned in the genome with respect to each other and relate the B73 maize physical map to the genetic maps by way of shared markers. As our view of the genome improves, the MaizeGDB Genome Browser’s representation of the genome will track that progress so that researchers will be able to make use of available data as it emerges.
Genome browser integration with the existing MaizeGDB resource
The MaizeGDB Genome Browser provides integrated views among genomic regions, genetic markers and sequences based upon the B73 sequence assembly. To enhance the capabilities of the Genome Browser and to better integrate it with the existing MaizeGDB resource, images of relevant genomic regions have been integrated into various pages at MaizeGDB [e.g. expressed sequence tag (EST)], genome survey sequence (GSS) and molecular marker pages] where each image is linked into the appropriate regions within the Genome Browser. In addition, the MaizeGDB BLAST (22) tool outputs were upgraded to show genomic locations of hits visually and a Locus Lookup Tool (21) that integrates genetic data with genomic views was created.
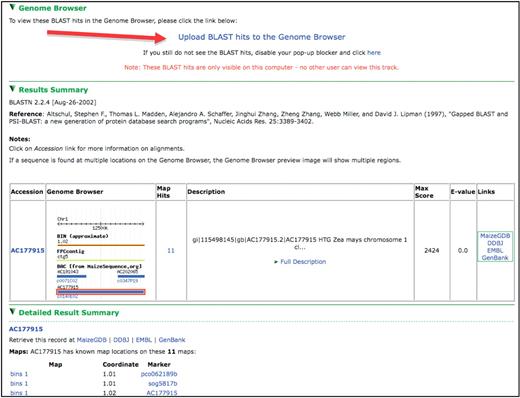
The MaizeGDB BLAST tool, which is locally run and updated monthly, is accessible from various locations within MaizeGDB, including the top of the Genome Browser itself (alongside various other MaizeGDB tools). BLAST searches can be performed against BACs, ESTs, GSSs, overgos and other maize nucleotide sequences. Each BLAST hit with known genomic coordinates shows a snapshot of the appropriate genomic context, the e-value of the hit and a short description of the genomic element (Figure 2). Researchers can click on the image to access that region in the MaizeGDB Genome Browser. Detailed information about the BLAST hit along with nucleotide-level alignments of sequences can be reached by scrolling down the results page.
A view from the BLAST results interface. Note the link toward the top of the page allowing BLAST hits to be uploaded as a track in the Genome Browser, results table showing a thumbnail of the hit’s genomic context and hit assignment to BACs via the molecular markers associated with the hit sequence.
To more precisely map a BLAST hit to the genome, functionality has been added to allow researchers to upload BLAST hits as a separate track to the Genome Browser. By clicking a button on the results page (red arrow in Figure 2), a BLAST track becomes exclusively available to the machine that launched the BLAST search.
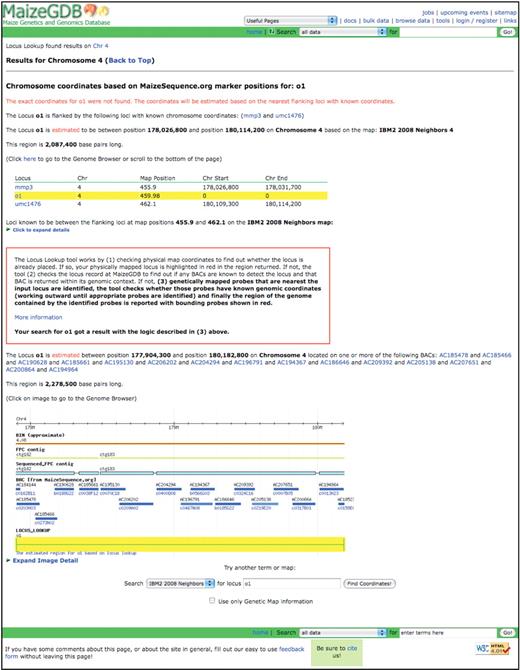
Maize is a species with a long genetic history, and over 1700 genetic maps are currently available via MaizeGDB. Because genomic coordinates are not available for all loci and some genetically mapped loci are not cloned, a mechanism for estimating the genomic location of such loci is needed for, e.g. researchers walking to genes. To meet this need, the Locus Lookup Tool (Figure 3) was implemented. The Locus Lookup Tool (available from the MaizeGDB front page, the Genome Browser and throughout MaizeGDB in other relevant locations) works by first checking physical map coordinates to find out whether the locus is already placed. If so, the physically mapped locus is highlighted in red in the appropriate genomic region. If not, the tool checks the locus record at MaizeGDB to find out if any sequenced BACs are known to detect the locus, and, if so, that BAC is returned within its genomic context. If not, genetically mapped probes that are nearest to the input locus are identified, the tool checks whether those probes have known genomic coordinates (working outward until appropriate probes are identified) and finally the region of the genome contained by the identified probes is reported with bounding probes shown in red.
The result page for the approximate genomic coordinates for the opaque endosperm1 (o1) locus when the Locus Lookup tool is used.
It should be noted well that even though a locus may physically map to a known location within a BAC, because most B73 BAC sequences currently are concatenated sequence fragments of unknown order and orientation, the locus position on each individual BAC sequence may be anywhere on the BAC associated with a physically mapped probe. For this reason, a conservative window showing the entire BAC(s) to which the probe(s) have been physically mapped is displayed.
Expanded structural and genetic map sets integrated with BACs and the B73 genome sequence
As mentioned previously, MaizeGDB is the central archive for maize maps and documentation. MaizeGDB map pages include views of mapped markers and genotyping scores, updated files for sequence-based markers with assigned map locations, GenBank (17) accession numbers and FASTA-formatted sequences as bulk downloads (see, e.g. the UMC 98 ‘sequence view’ for chromosome 1 at http://www.maizegdb.org/cgi-bin/displaymapwithaccessions.cgi?id = 143431). To facilitate linking genetic maps to the B73 genome sequence, BAC sequence accessions are imported monthly from GenBank, linked to MaizeGDB’s molecular marker data and assigned a bin coordinate. The BAC bin values are based on coordinates provided by the MaizeSequence.org FTP site (http://ftp.maizesequence.org/current/fpc_report.txt) or, in the case of some 900 B73 BACs sequenced by other groups, computed locally to be consistent with MaizeSequence.org coordinates. There are currently 61 000 loci with some genetic map information. These include the 15 568 sequenced BACs and 27 870 (mostly EST-based) markers resolved at the level of a BAC on the IBM2 FPC0507 maps (2). New recombination-based maps, described below, contribute a large number of sequenced markers that can be used for integrating genetic maps with the genome sequence.
Newly available data include the new generation ‘NAM’ (Nested Associate Mapping) maps from the Maize Diversity-Based Genomics project (22). These are fully documented, with genotype data provided pre-publication. They constitute 27 map sets, based on high-throughput single nucleotide polymorphism (SNP) genotyping of some 1100 markers for nearly 5000 recombinant inbred lines (RILs). The maps are sequence based and closely tied by related overgos to the B73 genome sequence (23). The mapping stocks were designed to support candidate gene discovery for agronomic traits using a nested association mapping strategy (24–26). The RILs are available from the Maize Genetics Cooperation–Stock Center [MGCSC (27)]. Documentation includes the genotype scoring for all the lines, sequence accessions submitted to GenBank, sequences of the allele-specific interrogation primers for B73 and Mo17 and allele descriptors for each relevant SNP with nomenclature based on the style developed for the maize TILLING (28) project (e.g. http://www.maizegdb.org/cgi-bin/displaylocusrecord.cgi?id = 978391). Links are provided to the source database as well as MGCSC.
Another addition is the Genetic 2008 map. It includes genes with experimentally confirmed gene products (1400) and/or phenotypes (380) that can be ordered to a resolution of 1–2 cM or better. The map was compiled manually from classical recombination data (29), along with sequence alignment to B73 BAC clones ordered by the maize FPC (13). New genes may be included by any cooperator directly by request.
The 2008 Neighbors Consensus maps incorporate the new genetic maps, including Genetic 2008. The current Neighbors maps include 15 904 loci, representing multiple marker technologies (6016 indels; 1388 SNPs; 1965 simple sequence repeats and 2153 overgos). Some 21% (3414) of these loci have been associated with the B73 genome BAC contigs by mappings based on sequence identity to markers/probes and/or high sequence similarity to markers/probes. This version of the Neighbors map differs from the previous iteration, IBM2 2005 (20), by including only markers ordered by recombination analysis and thus excludes most of the overgo and EST-based markers represented in the hybrid IBM2 FPC0507 map set (2).
Current endeavors
POPcorn—a PrOject Portal for corn
Maize researchers cannot easily leverage all available genetic and genomic data because the online locations of all resources are not easy to find and the sequence-indexed resources generated by individual projects must be searched independently. In addition, it is often the case that when a project’s funding period ends, the generated data are lost because they are not moved to long-term repositories: these once-funded project sites degrade over time and sometimes disappear entirely. The MaizeGDB team aims to overcome these challenges in collaboration with the community of maize researchers through POPcorn (PrOject Portal for corn; http://www.maizegdb.org/popcorn), a needs-driven resource and data pipeline. POPcorn offers (i) a centralized web-accessible resource to search and browse ongoing maize genomics projects and will make available (ii) a single, stand-alone tool that makes use of web services and minimal data warehousing to enable researchers to carry out sequence searches at one location that return matches for all participating projects’ related resources and (iii) a set of tools that enable collaborators to migrate their data to MaizeGDB at the project’s conclusion. A functional version of the POPcorn resource that serves as a portal to research projects has just been released. Sequence search capabilities and data upload tools are planned for release in early 2010.
Community-driven maize genome annotation
The BAC-by-BAC maize genome sequencing effort is nearly complete with > 17 000 Phase 1 BACs deposited in GenBank. The BACs are physically mapped (13). Although most BACs consists of several unordered sequence segments (or scaffolds) separated by unsequenced gaps, it is clear that sufficient sequence and map information is now available to derive gene models and assign them to classical genetic loci.
Maize stands out amongst the other plant genomes (e.g. Populus, Sorghum and Brachypodium) at similar annotation stages because of both the availability of rice and Arabidopsis as excellent model organisms to help the annotation effort and a large, well-organized research community that is ready to be engaged in the effort. Moreover, current annotation projects (including MaizeSequence.org, MAGI and PlantGDB) can be easily used as a springboard to complete community-driven annotation. For example, PlantGDB provides a daily updated table of new maize BACs from GenBank annotated with matching rice and Arabidopsis proteins (http://www.plantgdb.org/ZmGDB/DisplayGeneAnn.php). The table is sortable by location, rice gene product annotation or date and linked to genome context views with associated yrGATE community annotation tools. Thus, newly sequenced homologs of known rice or Arabidopsis genes are immediately accessible to the community for perusal and potential expert annotation.
In brief, the annotation process consists of: (i) a computational stage in which gene structures are predicted together with alignment evidence and (ii) subsequent rounds of manual annotation in which these models are improved. One set of computationally derived models for maize will be available from MaizeSequence.org (6). In addition, the EVidenceModeler (EVM) (31) meta-annotation system will be used to derive consensus gene structures. This program currently reconciles gene structures derived from a variety of computational gene finding tools and/or manual annotations according to a weight-based voting scheme, where weights are either set manually or learned using statistical training routines. Such systems have been shown to exhibit gene-finding competency commensurate with that of expert human annotators (32). As deployed for the maize community genome annotation project, EVM reconciles: gene structure predictions generated by the ab initio gene finders AUGUSTUS (33) and GeneMark.HMM (34), genes functionally supported by cDNA alignments as predicted by the GeneSeqer + MetWAMer system (35), as well as similarity data alignments generated by GenomeThreader [protein alignments (36)] and GeneSeqer [cDNA alignments (37)]. Elements in this panel of gene finders were selected largely based on their abilities to be retrained in a species-specific manner, enabling the generation of maize-specific parameter sets. It is anticipated that a more diverse collection of gene prediction software will be integrated into the pipeline over time. Once the maize genome has been computationally annotated using both de novo and comparative methods, instances of genes where cDNA and/or mRNA evidence are not congruent with the computationally derived annotation will be identified.
To facilitate community-driven maize genome annotation, the MaizeGDB and PlantGDB groups are working together. A brief overview of the annotation process is as follows. The gene models made available on MaizeSequence.org (6) and those derived from the EVM pipeline (described above) are evaluated for congruency with the most recent available data (EST and cDNA evidence that align to the gene). Next, a list of gene models that should be manually annotated is created using the xGDB GAEVAL (Gene Annotation EVALuation) system at PlantGDB (14) and made available to the community. Annotation project personnel contact researchers who may be working on particular genes in the list; the researcher annotates the gene directly using yrGATE, the xGDB genome annotation tool (38); and that annotation becomes available at both PlantGDB [via the maize xGDB browser called ZmGDB (39)] and MaizeGDB, pending approval by project personnel. Annotation project personnel will (i) conduct training in the use of yrGATE; (ii) alert members of the maize community to the need for curation of incongruent gene models that are of interest to them; (iii) encourage researchers to do the annotations (though this may seem minor, it is anticipated that this will require the majority of time and effort); and (iv) report the success of the project at conferences. The yrGATE training sessions will be conducted at the Annual Maize Genetics Conference and via three or more site visits per year to locations where many maize researchers are located.
It is anticipated that researchers may wish to annotate genes based upon: (i) genomic location; (ii) gene family membership; and (iii) involvement in particular biochemical pathways. To aid in identifying researchers and students who may wish to be involved in the genome annotation project, a query will be sent out to all maize cooperators asking for the contact information for at least one person per research group who will serve to annotate on behalf of the lab. Along with contact information, respondents will be asked for a list of locations/gene families/pathways of interest to the research group. Again, this effort in maize will benefit from the many resources now available for other model organisms and pan-species studies, e.g. http://www.kegg.com/, http://www.biocyc.org/, http://www.ebi.ac.uk/Tools/InterProScan/.
As outlined above, the MaizeGDB Team has chosen GBrowse to serve as the basis for the MaizeGDB Genome Browser. This is very helpful for working with PlantGDB’s xGDB and yrGATE systems given that GBrowse supports Distributed Annotation System (DAS; http://www.biodas.org) for data transfer and ZmGDB has DAS capabilities. PlantGDB’s ZmGDB resource will serve annotations as a ‘Community Annotation Track’ to the MaizeGDB Genome Browser via DAS, so researchers’ annotations will be available in real time. In addition, Ensembl (the genome browser software underlying MaizeSequence.org and Gramene) also supports DAS for sharing annotation data, which will make the generated gene models readily available for various plant database sites to pick up freely.
As with all other data generated by PlantGDB and MaizeGDB, all annotation efforts will be immediately and comprehensively available to the community, via web browsers, via DAS and by download. For example, two of the authors (M.E.S. and V.P.B.) recently identified 1665 non-redundant full-length maize genes on 1463 unique BACs that are highly conserved with Sorghum proteins (36), and which are served via DAS at http://sunx4600uno.gdcb.iastate.edu:9002/das/Zm_to_Sb with ProServer software (40). These data are proffered to the maize community as a high-quality gene set for use in training and assessing gene finding software tools, which can be accessed online at http://www.plantgdb.org/ZmGDB/DisplayZmToSb.php.
Outreach
Outreach continues to be an area of ongoing action at MaizeGDB. Since March 2007, tutorials have been taught at the University of Florida (Gainesville), the University of California (Berkeley), Stanford University (California), the USDA-ARS Plant Gene Expression Center (Albany, CA) and Iowa State University (Ames). In addition, MaizeGDB Team members are available for people to call or email with questions. Responding to feedback is a high priority, and the MaizeGDB team strives to be responsive to the suggestions and comments of maize research community members. In addition to live tutorials, online movie tutorials are also available with more currently in the making. These short movies demonstrate a specific topic of interest and are available online alongside other outreach materials at http://www.maizegdb.org/tutorial.
Funding
U.S. Department of Agriculture-Agricultural Research Service; National Science Foundation (grant numbers DBI 0743804 and 0606909). Funding for open access charge: U.S. Department of Agriculture-Agricultural Research Service.
Conflict of interest. None declared.
Acknowledgements
We thank the MGSC and MaizeSequence.org groups (especially Shiran Pasternak and Doreen Ware) for sharing the maize genome sequence data and their analyzed data sets with us and with the community prior to publication. We also thank the MaizeGDB Working Group (E. Buckler, K. Cone, M. Freeling, O. Hoekenga, A. Lamblin, K. McGinnis, L. Mueller, P. Schnable, M. Pop, T. Slezak, A. Sylvester, D. Ware, M. Sachs and V. Brendel) as well as the broad maize research community for their help and guidance. We also thank G. Davis, M. McMullen and E. Coe for advice on mappings.
References
Author notes
Present address: Michael E. Sparks, USDA-ARS Bovine Functional Genomics Laboratory, Beltsville, MD, USA