-
PDF
- Split View
-
Views
-
Cite
Cite
Fabio Rinaldi, Simon Clematide, Yael Garten, Michelle Whirl-Carrillo, Li Gong, Joan M. Hebert, Katrin Sangkuhl, Caroline F. Thorn, Teri E. Klein, Russ B. Altman, Using ODIN for a PharmGKB revalidation experiment, Database, Volume 2012, 2012, bas021, https://doi.org/10.1093/database/bas021
Close - Share Icon Share
Abstract
The need for efficient text-mining tools that support curation of the biomedical literature is ever increasing. In this article, we describe an experiment aimed at verifying whether a text-mining tool capable of extracting meaningful relationships among domain entities can be successfully integrated into the curation workflow of a major biological database. We evaluate in particular (i) the usability of the system's interface, as perceived by users, and (ii) the correlation of the ranking of interactions, as provided by the text-mining system, with the choices of the curators.
Introduction
The increasing quantity of available biomedical data pose a challenge to life scientist wishing to explore a particular research problem. Although emerging bioinformatics services enable structured access to increasingly complex datasets, it is often the case that the primary data is only available in the published literature, and needs to be extracted and stored in a standardized format before it can be leveraged upon (1). This is the main motivation behind biomedical curation activities: ‘to help the Life Sciences community to make sense of all the data that is accumulating’ (2).
Although human curation offers the best guarantee of high-quality results, it suffers from severe bottlenecks that have long been recognized in the curation community. The most pressing problem is that of efficiency of the process: despite the fact that typically several databases attempt to focus on a particular type of biological data, and often collaborate at least sufficiently to prevent duplication of effort and ensure compatibility of resulting data formats, it is impossible for human curators to keep up with the growing pace of publication.
Nobody will ever be able to manually annotate all the macromolecular biological entities that exist on this planet, and consequently automatization is the only solution. (2)
On the other hand automated text-mining tools cannot offer sufficient reliability to be employed indiscriminately without human supervision of the results that they deliver. Therefore, the ideal solution is to combine the best capabilities of automated systems with human supervision by highly qualified domain experts.
In this article, we describe recent experiments aimed at assessing the potential contribution of a specific curation tool (ODIN) to the curation process of a well-known database (PharmGKB). In the rest of this section we briefly describe both PharmGKB and ODIN.
The Pharmacogenomics Knowledge Base (PharmGKB) is a publicly available online worldwide resource (www.pharmgkb.org) (3,4). The mission of PharmGKB is to collect, encode and disseminate knowledge about the impact of human genetic variations on drug responses, contributing to the drive towards personalized medicine for better therapeutics. PharmGKB is an NIH-funded resource, which over the past 11 years has maintained a very high-quality manually curated knowledge base of pharmacogenomics facts, curated by a team of PhD and Masters level scientists.
One of the many tasks of the PharmGKB curators is to review past and current literature and add any relevant pharmacogenetic or genomic articles to the PharmGKB database. The curators identify relevant journal articles, largely selected from a set of about 20 journals that are followed, which include major pharmacogenomic journals and publications published by the PGRN (Pharmacogenomics Research Network). They then read the abstract or full text if necessary, and populate the knowledge base with information about the genes, drugs and phenotypes discussed. In the past, this information was gathered in Microsoft Excel spreadsheets and uploaded to the database. Today, the information is entered through a web-based graphical user interface (GUI) developed in-house to fit the curators' needs, and captures data that is far more structured than in the past, such as population characteristics of the study group described in an article, and P-values of associations found between genetic variants and drug response. Curators can manually enter drug and gene terms for each article using auto-complete fields that draw on PharmGKB's standardized vocabularies. Additionally, in the past, in each article curators captured the entities discussed in the form of a list of genes, drugs and phenotypes, which did not enable users to presume binary relationships between a single gene and a single drug in a PharmGKB Literature Annotation. Today, the relationships between entities are binary, such that for example a gene–drug relationship is explicitly captured, including some degree of specification regarding the type of interaction (‘is associated with’, ‘inhibits’, etc.). A detailed description of the types of annotations in PharmGKB has been published previously (5).
The current curator GUI assists the curators in their process using basic text mining by suggesting entities found in the article, but does not pre-populate fields or highlight any information found within the article text. The PharmGKB team is currently working on developing Natural Language Processing and machine learning methods to aid in the future in tasks such as document retrieval and information extraction.
The OntoGene group at the University of Zurich has developed advanced solutions for several text-mining tasks based upon advanced natural language processing technologies, which have been proven to be state-of-the-art by participation in several competitive evaluations (6–9). The OntoGene text-mining system is based on a standard NLP pipeline, composed of efficient modules for sentence splitting, tokenization, entity recognition, syntactic chunking and dependency parsing. Its entity recognition component has been shown in the recent CALBC shared evaluation (9) to be highly efficient and capable of delivering competitive results for several entity categories. Its relation mining component has been used in the BioCreative 2009 evaluation to deliver the best results for the identification of protein–protein interactions (7). [A more detailed description of the architecture of the OntoGene text-mining system is beyond the scope of this article, for further details the interested reader is invited to consult the following publications (6,7). Specific adaptations that were carried out for the PharmGKB task are described in separate forthcoming publications (10,11)].
The results of the OntoGene text-mining system are made accessible through a curation system called ODIN (OntoGene Document INspector; this tool is not connected in any way with the recently introduced commercial text analytics system called OdinText.) which allows a user to dynamically inspect the results of their text-mining pipeline. A previous version of ODIN was used for participation in the ‘interactive curation’ task (IAT) of the BioCreative III competition (12). This was an informal task without a quantitative evaluation of the participating systems. However, the curators who used the system commented extremely positively on its usability for a practical curation task.
More recently, the OntoGene group created a version of ODIN that allows inspection of abstracts automatically annotated with PharmGKB entities [the annotation is performed using the OntoGene pipeline (http://www.ontogene.org/pharmgkb/)]. Users can access either preprocessed documents, or enter any PubMed identifier and have the corresponding abstract processed ‘on the fly’. For the documents already in PharmGKB it is also possible to inspect the gold standard and compare the results of the system against the gold standard. The curator can inspect all entities annotated by the system, and easily modify them if needed (removing false positives with a simple click, or adding missed terms if necessary). The modified documents can be sent back for reprocessing if desired, obtaining therefore modified candidate interactions. The user can also inspect the set of candidate interactions generated by the system, and act upon them just as on entities, i.e. confirm those that are correct, remove those that are incorrect. Candidate interactions are presented ordered according to the score that has been assigned to them by the text-mining system, therefore the curator can choose to work with only a small set of highly ranked candidates, ignoring all the rest.
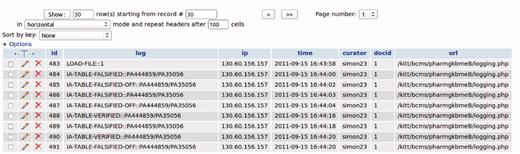
ODIN, which is based on a client–server architecture, maintains a log of the interaction with the curator, which could be used for later revision by a supervisor or for reversing some specific annotation decisions. At the end of a session the modified document and its annotations are sent back to the server, together with the log, for permanent storage, and can be accessed again at the next session, which could take place on a different remote client. Additionally, the curator can choose to export the annotations to a local file in a simplified format (e.g. comma-separated values).
Related work
Automated tools have the potential to support the curation process in several phases. First of all, text-mining tools can provide a help in the initial triage stage in order to decide which papers should be inspected by the expert curators. Text classification tools are nowadays capable of reliably processing large sets of articles in order to score them and provide a ranked list of candidate papers, which can then be used to prevent inspection of less promising articles. This process is typically based on machine learning tools that can distinguish interesting and less interesting articles on the basis of similarities with previously classified articles.
During inspection of individual articles, it can be very helpful for curators to use a system capable of locating the entities of interest within the article, and disambiguate them as reliably as possible. This process is based on named entity recognition tools, which recently have made considerable progress and are now capable of recognizing several types of biomedical entities with great reliability. For example, recent results in the BioCreative competition (13) have shown that several systems are capable of recognizing and disambiguating gene names with F-scores above 80%. Databases that are entity-focused can immediately profit from such tools, as the curators will be able to manually filter the candidates suggested by the system at greater speed, compared with a manual extraction from the paper, which would involve (i) spotting the mentions in the paper, (ii) decide which database entities are actually intended.
The next major challenge for the introduction of text-mining systems within curation workflows is the automated detection of relations, which is relevant for several databases. Tools that can reliably detect entity interactions are in general much less efficient than named entity recognition tools due to the much greater complexity of the problem. In order to produce candidate interactions a tool needs first to identify the entities correctly. Given that some errors are inherent in this process, generation of candidate entity pairs will inevitably result in compounding that error, leading to lower performance. Contextual clues that can help to identify an interaction candidate are typically very sparse, making difficult to apply machine learning techniques.
Nonetheless, much progress has been achieved recently, as results in the BioCreative II (14) and II.5 (15) competitions show, and therefore it is now appropriate to start practical experimentation through collaborations between developers of text-mining solutions and database groups as potential users. Although immediate integration in the curation workflow might not be the goal, these joint experiment help both groups in deciding how to improve their activities.
Text-mining developers will receive feedback on the quality of their systems and gain an understanding of the specific needs of the curators [(12) stresses the ‘importance of understanding the biocurator's curation workflow’], and curation groups will gain a better understanding of the current potential of technologies, which are now still experimental, but might soon become mainstream, and thus be able to choose the optimal point for integration in their workflows. Additionally, the feedback provided to text-mining developers will render future systems more usable in practical applications.
The need to pair developers with curators has been recognized by the organizers of the BioCreative competition. A new experimental task (IAT) dedicated to the evaluation of interactive curation environments was introduced in the last edition (12). Although the specific task chosen for the experimentation was an entity recognition task, several of the conclusions reached through analysis of participating systems are applicable to all types of interactive curation environments. Addressing usability of text-mining systems is a novel aspect of this task. ‘Usability … enables the users to find, interact with, share, compare and manipulate important information more effectively and efficiently’ (12).
Although fully unsupervised extraction of information from the literature is, for some time at least, unrealistic, text-mining tools are already sufficiently reliable to be used to provide hints to the curators, in order to speed up their activities. Such a help is sorely needed, as it is already clear that manual curation cannot keep up with the rate of data generation (16). Curatorial work done with the assistance of a text-mining system has already been shown to be much more efficient than when done by human readers without support (17). The authors of this study state that: ‘For biologists, an automated system with high recall and even moderate precision … confers a great advantage over skimming text by eye.’ Examples of well-known text-mining solutions are iHOP (18) and ChilliBot (19). Among the systems developed to support the curation process, one of the most interesting is (20). They use a manually annotated corpus (gold standard) to simulate an assisted curation environment, where the curators are given either gold standard data or the output of an (imperfect) NLP pipeline. (20,21) presents a system developed for the curators of FlyBase, a database for drosophila genetics and molecular biology. Although the document analysis is based on a conventional NLP pipeline, including the dependency parser RASP (23), the curator's interface has been developed in strict collaboration with the end-users. (24) discuss how well the performance of a text-mining system (in their case tailored to identify mentions of protein mutations), when evaluated with conventional techniques, translates into real utility of the system for a curation task. Textpresso is another well-known text-mining system that is characterized by the usage of ontological categories of biological concepts (17,25), as well as by processing full papers. A variant of Textpresso (Pharmspresso) has been used for automatic annotation of pharmacogenomic literature for PharmGKB, but was never integrated with the manual curation process (26).
Methods
Although the full OntoGene pipeline can deliver reliably a ranked list of candidate interactions, which can then be used by curators as prompts for annotation of novel articles, the experiment described in this article centered upon the validation of existing relations from PharmGKB. Revalidation of existing data is a common practice of several biological databases, for example (2) mentions several steps of re-annotation for Swiss-Prot, one of the most well-known and authoritative databases.
The main aim of the experiment was therefore to evaluate the usability of the interface, rather than the capabilities of the underlying text-mining tools. As (12) points out, an evaluation task must be chosen to be feasible in a given time frame, considering both the time needed by developers to adapt the existing text-mining system to the specific needs of the applications and the time available to curators for the verification of the results.
We started by considering the set of articles already curated by PharmGKB, processing them with the OntoGene relation extraction system. We then automatically compared for each article the results of the relation mining system with the manually extracted interactions, and computed the common subset. In general, we would expect the text-mining system to deliver a larger set of interactions than those manually curated (ideally covering all of them). In practice, since in this experiment only abstracts rather than full text were used, this was true only in 3059 articles out of 5378. All the remaining articles contain at least one ‘false negative’, i.e. a relation that was not detected by the text-mining system. (This could be due to several factors, for example an interaction that is mentioned only in the full text and not in the abstract will be obviously impossible for the system to detect with the current settings. Another possible source of false negatives is due to the way the PharmGKB data was created, i.e. for each paper, a list of genes and drugs discussed was kept, and in some cases interactions among those entities were simply hypothesized and not actually verified. Therefore, some of the interactions in the PharmGKB data are not expected to be true positives). If full articles had been processed, we would have expected the number of false negatives to be much lower. Full articles, however, are difficult to process for several reasons, most of which have little to do with text mining, such as widely different formats, or copyright restrictions, which in some cases explicitly prohibit text-mining applications.
We decided for this experiment to use only articles from the set where all interactions were found by the OntoGene pipeline. The aims of the experiment were the following: (i) evaluate the usability of the interface for revalidation of PharmGKB relationships, (ii) estimate whether the ranking of interactions provided by the text-mining system correlates well with decisions taken by the curators. The previously developed ODIN system was adapted to the needs of PharmGKB, on the basis of a close interaction with the curators. During development the following recommendations by (12) (adapted to the specific needs of our application) were taken into consideration:
support for interactive disambiguation of domain entities;
an editable list of candidate entities or interactions;
a view of the document correlated with the candidate interactions (when an interaction is selected, the corresponding entities are highlighted);
ability to sort the results according to different criteria;
ability to collect event and timing information at the session level; and
ability to export the results in a suitable format (e.g. CSV).
The ODIN system allows the user to verify and modify every single annotation provided by the system, at the entity level as well as at the interaction level (a). It provides a ranked list of candidate interactions (b), which additionally can be sorted by the user according to different criteria (d). The interface is structured around three panels: term editing panel, document panel and results panel. The term editing panel (not shown in the pictures in this article) can be used for (a). The document panel and results panel are actively connected, in that selection of items from the results panel will result in their visualization (highlighting) in the document panel. Every action of the user is stored in a log that is stored at the server level (e) with timing information (Figure 2). Finally, the results can be exported as a CSV file (f) and additional formats can easily be added upon request.
The close interaction among curators and system developers allowed the latter to implement a number of suggestions that made the usage of the system more effective. For example, in the initial demonstration the entities participating in an interaction were represented only by the PharmGKB identifier of the participating entities. The curators pointed out that it was not immediately obvious to them which entity was referred to by the identifier without consulting the database (that the ODIN system allows by simple click on the identifier, see Figure 1), so the ‘reference name’ of the entity was added. This, however, made the table of interactions more cluttered, and the curators pointed to the fact that they might optionally want to remove some information from the table (such as the entity identifiers). The developers therefore modified the interface to allow precisely this type of modification directly by the user (i.e. selection of which fields they want to be displayed), see Figure 3. Some of the information hidden in this way could further be displayed as unobtrusive tooltip windows on mouseover by the user, another option that was added upon suggestion by the curators. (1) stresses the importance of being able ‘to hide fields of negligible value to the curators thereby distracting their attention unnecessarily’.
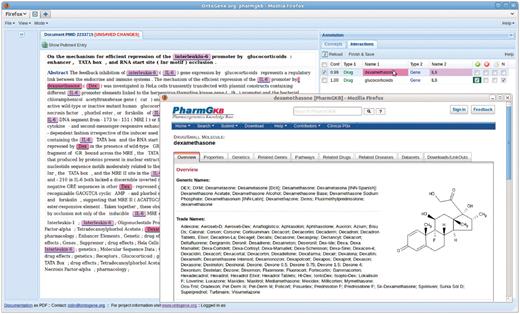
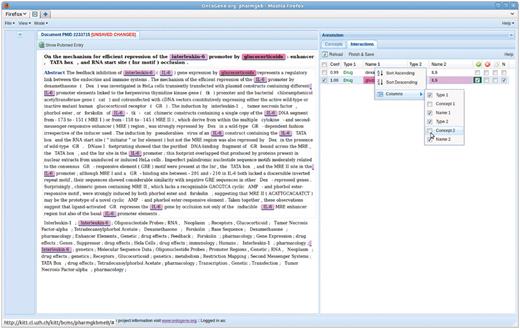
Another example of the fruitfullness of the interaction between developers and curators is the addition of different types of confirmation boxes for an interaction. Instead of a simple confirm/reject choice, the maintainers of the database suggested the need for a more fine-grained choice. In particular, they wanted to be able to confirm ‘negative’ interactions, i.e. interactions that are stated in the paper as NOT to hold under the conditions investigated. Another wish was to be able to state that the abstract inspected for the experiment did not provide sufficient information to either confirm or reject the proposed relation. Figures 1, 3, and 4 show these options as four tick boxes in the top right corner of the picture, which correspond to ‘confirm relation’, ‘reject’, ‘needs full text’, ‘negative relation confirmed’.
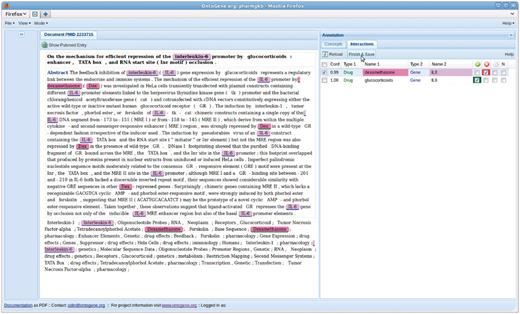
Entities which participate in the selected interaction are highlighted in the document panel.
After a preliminary test phase on a few selected sample articles, which allowed the curators to gather some familiarity with ODIN, and the developers to fix the remaining issues, the validation experiment could start. A set of 125 articles was selected from the 3059 articles where the OntoGene pipeline could detect all of the relations originally annotated by PharmGKB. The selection was based on a randomized stratified sampling process, in order to generate a distribution of relations per article that would be roughly equivalent to the distribution in the whole set. This set was split into five sets of 25 articles each, which were then randomly assigned to PharmGKB curators. This sampling lead to the following distribution of articles per curator: 8 articles with 2 relations, 9 with 3 relations, 3 with 4 relations, 3 with 5 relations, 1 with 6–7 relations, 1 with 8–9 relations, 1 with 10–20 relations.
The data sets were at all stages identified only by a symbolic reference (A, B, C, D, E), which was randomly assigned to the curators (and known only to each of them), in order to ensure anonymity. This was done to avoid generating the impression that the result of the experiment could be used to evaluate individual performance. The full cooperation of the curators is of utmost importance to guarantee unskewed results, therefore we took care to prevent the possibility of identification. Curators were then asked to inspect the articles in the assigned set with the ODIN system and then use it to validate the interactions. During this process all of their actions were logged using the symbolic reference (that they had to enter into the system at the beginning of the process) as an identifier. The resulting validation decisions are automatically saved by the system and transferred to a server, together with detailed logs of the activity. This data set forms the basis of the evaluation presented in the next section.
At the end of the curation experiment we asked the curators to fill a questionnaire that was partly modeled on the questions used in the BCIII IAT Task (12). The feedback received through this survey is discussed at the end of the next section.
Evaluation
As explained in the previous section, the experiments described in this article were centered on the revalidation of relations already stored in PharmGKB. In order to evaluate the correlation of the rankings provided by the text-mining system with curator's validation decisions, we were limited to use only articles for which all PharmGKB interactions could be detected by the text-mining system. Since only abstracts were used for automatic processing, only about 56% of PharmGKB curated articles (3059) could be taken into consideration.
We also observed that articles that contained a single curated relation would not be particularly interesting for this experiment, since it can be presumed that the vast majority of these cases are correct, and in any case there is no ranking to evaluate. Additionally, articles containing more than 20 interactions were also excluded, because there was a very limited number of them, and they would require too much time for revalidation. Excluding these cases, we were left with a set of 1407 articles. Out of this set, we selected by stratified random sampling five sets of 25 articles each, as described in the previous section.
In the rest of this section, we describe in detail the results of our experiments through descriptive statistics computed from the logs of the interactions. This is followed by a qualitative analysis of the final survey.
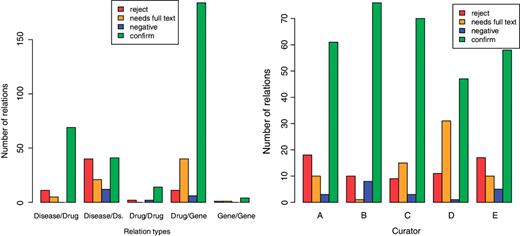
The curators could take for each relation one of four decisions: ‘confirm’ (the abstract supports the interactions), reject (there is no support in the abstract for the interaction), ‘negative’ (the abstract states that the mentioned entities DO NOT interact), ‘needs full text’ (there is no sufficient information in the abstract to decide either way). The pie chart on the left of Figure 5 shows the total distribution of these decision across all articles. Nearly 3/4 of the relations were confirmed as positive. However, this distribution appears to be strongly dependent on the type of the entities participating in a relationship. The distribution of such decision by relation type is shown on the left of Figure 6. The relationship Drug/Gene has been chosen to be the main focus of future revalidation work and the results show that this type of interaction has a relatively low rejection rate. The distribution by curator is shown on the right of the same figure.
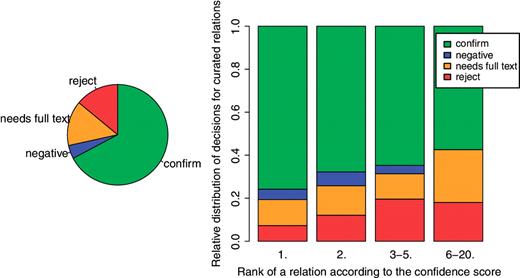
One of the aims of the experiment was to verify how the ranking of interactions produced by the text-mining system correlates with validation decisions by the curators. The bar chart on the right of Figure 5 shows a clear positive correlation at least for the best ranked cases (ranks 1–5). The proportion of interactions that the curators confirm as positive is greater at rank 1 and gradually decreases. At higher ranks there is no visible correlation, but this is partially due to the sparsity of data (in general there are fewer articles that have 5 interactions, and very few that have more than 20) (Among the articles selected for the experiment, 30.06% have 2 interactions, 37.81% 3 interactions, 6.39% 4 interactions, 11.02% 5, 5.90% 6 or 7, 4.33% 8 or 9 and 4.48% 10–20.).
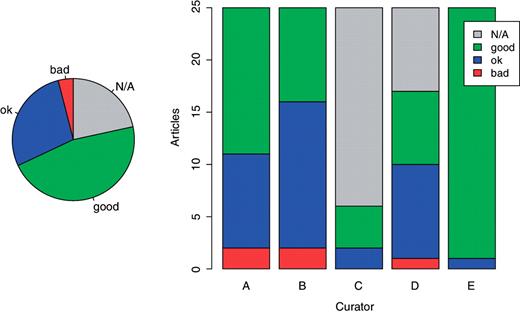
After validating all relations in each document, the curators were asked to express their opinion about the quality of concept identification (provided by the text-mining system) for that particular document. They could only chose among three values (bad, ok, good). However, since this comment was not mandatory, in about 1/4 of articles such judgments are missing. Figure 7 on the left shows the totals, and on the right distributed per curator. These values represent the perceived quality of concept identifications, i.e. the subjective judgment of the curators about the correctness of the entities suggested by the system.
In our experiment we also measured (through the logs) the exact time span between opening of a document and saving it after completing the validation of its interactions. This time was then divided by the number of interactions that had to be validated in each specific document. The average time needed for the validation of each interaction varies strongly among different curators, from 15 s up to 122 s. The box-and-whisker plots in Figure 8 illustrate the distribution of the mean time (in seconds) used for the curation of all relations of an article. The graph on the left shows these timings in relation to the curator's subjective judgment about the quality of concept recognition in the article, the graph on the right shows the timings per curator. The bottom whisker gives the minimum mean time, the top whisker gives the maximum mean time. Whiskers are shortened as usual to a length of 1.5× the box length and possible outliers are plotted separately with points. The bottom line of box is the first quantile, the upper line of box is the third quantile. The median is marked by the strong black line in the box.
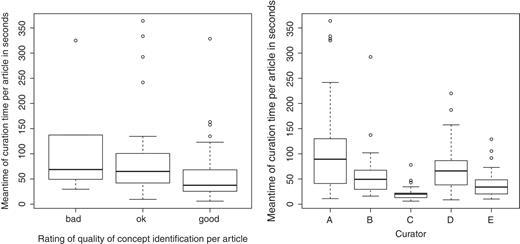
Box-and-whisker plots illustrating curation time (on the left according to the decision taken, on the right per curator).
An interesting research question is to evaluate whether the quality of a text-mining solution has a correlation with the time necessary for the curation of a specific article. The left graph in Figure 8 clearly shows such a correlation for our experiment: articles where concept identification was regarded by curators as ‘good’ required a much lower curation time than for the other two categories. It should be noticed, however, that the category ‘bad’ was selected in a too small number of cases, and therefore results in this category might not be very informative.
As mentioned in the previous section, at the end of the experiment the curators were asked to fill in a brief questionnaire in order to collect subjective feedback about their experience with the curation environment. One of the questionnaires was not returned. Below we list the questions that were asked and the feedback received.
Q1 Do you consider the system easy and intuitive to use?
not intuitive at all
partly intuitive (25%)
mostly intuitive (75%)
very intuitive
Q2 Do you consider the organization of the panels to be practical?
not practical at all
partly practical
mostly practical (100%)
very practical
Q3 What aspects of the system are most appealing to you?
Having the abstract to the left of the terms makes sense to me. It did help in that the entities in a proposed relationship were listed already so matched a PharmGKB term and were spelled correctly. That saves a bit of time.
Speeds things up because relationships are already entered just need to verify them
Highlighting the genes and drugs in the abstract, especially matching gene synonyms to gene symbol.
Q4 What problems/limitations of the system did you notice?
I would like to see the mapping of terms that are different in the abstract versus displayed gene names or diseases in the panel. I would like if just the gene and drug would be highlighted in the sentences used by the program to define the relationship not every mention of the gene and drug in the abstract. At the moment too many objects are highlighted.
Whenever I checked the green checkbox, it would turn on the highlighting for that row(I did not have that experience with the other choices).
I think it missed some relationships.
Sometimes needs to resort to full text. Some gene/drug relationships may not be identified through the system but are true relationships. Would be more valuable to extract the types of relationships between the concepts (eg. metabolize, transport, inhibit, induce etc)
Q5 Please mention any aspect of the system that did not appeal to you, or suggestions for changes.
I would have liked an option to turn off the underlining in the abstract. I find highlighting and underlining to be distracting and to clutter up the thing Im trying to read; I do not find them helpful. Maybe there was an option to do that and I simply did not notice it.
It would be helpful to have a place to add free text note to each relationship
Would be more valuable to extract the types of relationships between the concepts (eg. metabolize, transport, inhibit, induce, treat etc), also would be nice to extract genetic variations as another type of concept.
Q6 Was the system helpful in performing the validation task?
not helpful at all
partly helpful
mostly helpful (75%)
very helpful (25%)
Q7 Would you consider using a similar system for your regular curation task?
no
probably not
probably yes (75%)
yes (25%)
Q8 Do you agree that a similar system could increase the efficiency of the manual curation process?
no
probably not
probably yes (75%)
yes (25%)
Discussion
Usability issues are crucial for the acceptance of any specific IT tool by the end users. In the case of biomedical curation, it is essential that text-mining results are delivered to the curator in a transparent fashion, without the need of dealing with system technicalities, in order to prevent cognitive overload. The tool should not disrupt the ‘rhythm, flow of thinking and mental modeling process’ of the users, otherwise even minor problems with the interface could turn into major disruptions (1).
Ideally the user should be put in a situation where he/she can make a quick but well-motivated decision based on the information provided by the system. According to (1) the output of a text-mining system should respect the following five criteria in order to be really helpful in the curation process:
Relevance: a connection to the disease of interest or to synonyms, homologs of interest.
Valid: not likely to be a false positive (has some statistic of significance associated with it).
Credible: trustworthy methods generated the evidence plus numerous lines of evidence, number of research publications, convincing public metadata.
Plausible: function, location/structure, interaction type, biological process, and cellular component suggest an explanatory story.
Manageable: enough interactions for promising insights but not so many as to be overwhelming (roughly between 10 and 50).
Another point mentioned by the same author is that it would be very helpful if the tool could present an interaction type: ‘Interaction type was crucial for scientists' judgments about whether results might help construct a plausible explanatory story.’ This is a wish that has also been expressed by the curators in the experiment described in this article.
On the basis of the final survey, it appears that the users appreciate the comfort and support that ODIN gives them. They consider it as helpful in several ways. According to the qualitative feedback provided in the survey, visual highlighting depends on personal preferences, and therefore users should be given the possibility to customize some additional aspects of the interface. Several curators mentioned that they would like to be able to add more specific information at the level of individual relationships, for example by means of a free text comment. Particularly useful would be to add to each relation an indication of its type as mentioned in the document (inhibition, activation, etc.). [The PharmGKB group is separately researching similar issues (27,28)]. If the system could provide hints in this direction, this would be a very helpful feature. The OntoGene text-mining system is capable of extracting this kind of interaction type indicators, however this feature was not used in the experiment as the developers assumed it would not be needed. This is another example that shows the importance of close collaboration between system developers and database curators.
Conclusion and future work
The experiment described in this article aims primarily at verifying the usability of the ODIN system in the context of curation of the PharmGKB database. The initial assumption was that we could separate an evaluation of the interface from an evaluation of the underlying text-mining system by asking curators to perform a revalidation task rather than a novel extraction task. The revalidation task consists in using the ODIN functionalities to quickly check the correctness of interactions already stored in PharmGKB.
The results of the experiment confirm the initial assumptions: (i) the ODIN system offers a comfortable environment for relation validation that can considerably speed up this particular curation task (ii) the positive validation decisions are strongly correlated with the rankings provided by the text-mining system.
As a next step, we intend to verify the quality of the interaction mining component on novel unseen articles but using the same interface. Since the curators at this stage are already familiar with the interface, we will be able to verify how the results delivered by the text-mining system can actually improve their effectiveness in annotating interactions, without being impaired by an unfamiliar interface.
Funding
The OntoGene group (F.R. and S.C.) is supported by the Swiss National Science Foundation (grant 100014-118396/1) and Novartis Pharma AG, NIBR-IT, Text Mining Services, CH-4002, Basel, Switzerland. The PharmGKB group (all other authors) is supported by National Institute of Health/National Institute of General Medical Sciences (R24 GM61374) and National Library of Medicine (contract HHSN276201000025C).
Conflict of interest. None declared.



