-
PDF
- Split View
-
Views
-
Cite
Cite
John Meinken, Gary Walker, Chester R. Cooper, Xiang Jia Min, MetazSecKB: the human and animal secretome and subcellular proteome knowledgebase, Database, Volume 2015, 2015, bav077, https://doi.org/10.1093/database/bav077
Close - Share Icon Share
Abstract
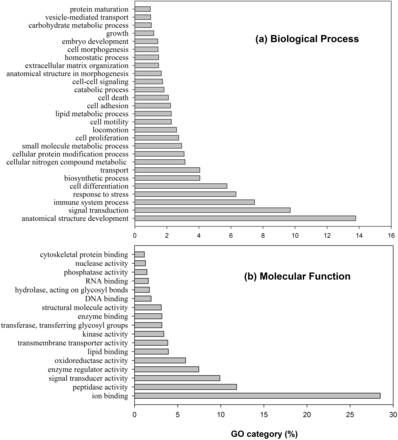
The subcellular location of a protein is a key factor in determining the molecular function of the protein in an organism. MetazSecKB is a secretome and subcellular proteome knowledgebase specifically designed for metazoan, i.e. human and animals. The protein sequence data, consisting of over 4 million entries with 121 species having a complete proteome, were retrieved from UniProtKB. Protein subcellular locations including secreted and 15 other subcellular locations were assigned based on either curated experimental evidence or prediction using seven computational tools. The protein or subcellular proteome data can be searched and downloaded using several different types of identifiers, gene name or keyword(s), and species. BLAST search and community annotation of subcellular locations are also supported. Our primary analysis revealed that the proteome sizes, secretome sizes and other subcellular proteome sizes vary tremendously in different animal species. The proportions of secretomes vary from 3 to 22% (average 8%) in metazoa species . The proportions of other major subcellular proteomes ranged approximately 21–43% (average 31%) in cytoplasm, 20–37% (average 30%) in nucleus, 3–19% (average 12%) as plasma membrane proteins and 3–9% (average 6%) in mitochondria. We also compared the protein families in secretomes of different primates. The Gene Ontology and protein family domain analysis of human secreted proteins revealed that these proteins play important roles in regulation of human structure development, signal transduction, immune systems and many other biological processes.
Database URL:http://proteomics.ysu.edu/secretomes/animal/index.php
Introduction
Secreted proteins play important roles in the development of multicellular organisms, serving as signal molecules, extracellular enzymes and structural matrix. The first sequenced protein, human insulin, was actually a secreted protein. Human secreted proteins have potential to be used as biomarkers for the diagnosis of diseases ( 1 ). The term ‘secretome’ was first used by Tjalsma et al . ( 2 ) to include all proteins that are synthesized and processed by the secretary pathway and proteins located in the secretion machinery. However, the term recently was limited to include only the set of secreted or extracellular proteins in a species ( 3 , 4 ). The secretome plays a central role in creating an extracellular environment that allows for physiological coordination and maintaining the homeostatic conditions that support cellular life and thus the organism.
Because of biomedical importance, secretome identification and analysis have been carried out in a number of human and animal cells or tissues including human arterial smooth muscle cells ( 5 ), human oligodendrocytes ( 6 ), human mesenchymal stem cells ( 7 ), human and mouse preimplantation embryos ( 8 ), primary human adipocytes during insulin resistance ( 9 ), rat adipose tissues ( 10 ), 23 cancer cell lines ( 11 ), and different types of human primary cell cultures and human body fluids including plasma, cerebrospinal fluid and urine ( 12 ). In addition to experimental characterization of human secretomes in various cell types, proteome-wide computational prediction of secretomes has been performed in mouse ( 13 ), human, pufferfish, pigs, and zebrafish ( 14 , 15 ). A secreted protein database was developed for human, rat and mouse, but unfortunately this database has not been updated since 2006 ( http://spd.cbi.pku.edu.cn/ ) ( 16 ), and another database, LOCATE, describing the membrane organization and subcellular location including secreted proteins was developed for mouse and human only ( http://locate.imb.uq.edu.au/ ) ( 17 ). However, as the complete genome sequencing projects have generated many complete proteome data in animal species, a database having information for computational prediction and curated information of secretomes and other subcellular proteomes in these species would provide a useful resource for both searching an individual protein subcellular location and performing proteome-wide comparative analysis.
In this work, we describe MetazSecKB, the Metazoan, i.e. human and animals, Secretome and Subcellular Proteome Knowledgebase. MetazSecKB is constructed with all available human and animal protein sequences by combining curated subcellular information and predicted information, with a well tested computational protocol, on secretomes and other subcellular proteomes of 15 subcellular locations. This knowledgebase is expected to serve as a central portal for providing information on metazoan protein subcellular locations for biological and medical researchers interested in protein biology.
Data collection and database implementation
Data collection
The protein sequences for the kingdom Animalia, also called Metazoa, were retrieved from the UniProtKB/Swiss-Prot dataset and the UniProtKB/TrEMBL dataset (release 2014_01) ( http://www.uniprot.org/downloads ). The UniProtKB/Swiss-Prot dataset contains manually annotated and reviewed protein sequences with information extracted from literature of experimental results and curator-evaluated computational analysis ( 18 ). The UniProtKB/TrEMBL dataset contains computationally analysed protein sequences. The combined metazoan dataset consisted of a total of 4 080 818 protein entries with 103 088 and 3 977 730 entries from the UniProtKB/Swiss-Prot dataset and the UniProtKB/TrEMBL dataset, respectively. The identifier mapping data including UniProt accession number (AC), UniProt ID, RefSeq accession number and gi number were retrieved from the UniProt ID mapping data file.
Protein subcellular localization prediction
We have previously evaluated several computational tools for predicting classic secreted proteins, i.e. proteins having a secretory signal peptide at the N-terminus ( 19 ) (Min 2010). These tools were chosen because they have relatively high prediction accuracy and are available as standalone tools for local processing of large datasets. The protein sequences were processed using the following programs: SignalP (version 3.0 and 4.0) ( 20 , 21 ), Phobius ( 22 ), WoLF PSORT ( 23 ) and TargetP ( 24 ) for secretory signal peptide and subcellular location prediction. TMHMM (version 2.0) was used to identify proteins having transmembrane domains ( 25 ) and Scan-Prosite (called PS-Scan in standalone version) ( http://www.expasy.org/tools/scanprosite/ ) was used to scan endoplasmic reticulum (ER) targeting sequence (Prosite: PS00014) ( 26 , 27 ). Proteins having one or more membrane domains, but not located within the N-terminus (the first 70 amino acids), were predicted as membrane proteins by TMHMM. The tools mentioned above were installed on a local Linux system for data processing. The commands for running these tools were summarized by Lum and Min ( 28 ). Protein sequences predicted to have a signal peptide by SignalP (version 3) were further processed using FragAnchor webserver to identify the glycosylphosphatidyinositol (GPI) anchors ( http://navet.ics.hawaii.edu/∼fraganchor/NNHMM/NNHMM.html ) ( 29 ). These tools have been used for processing fungal and plant protein sequences in construction of FunSecKB ( 3 ), FunSecKB2 ( 4 ) and PlantSecKB ( 30 ). However, based on our previous evaluations, the detailed methods were slightly different for assigning secretomes in different kingdoms of eukaryotes ( 19 ).
The metazoan protein subcellular locations are classified into the following categories: secreted proteins, mitochondrial (membrane or non-membrane), ER (membrane or lumen), cytosol (cytoplasm), cytoskeleton, Golgi apparatus (membrane or lumen), nuclear (membrane or non-membrane), vacuolar (membrane or non-membrane), lysosome, peroxisome, plasma membrane, other membrane and GPI-anchored proteins. For assigning a protein subcellular location, the UniProtKB subcellular annotation information was considered prior to using prediction information. For proteins not having annotated subcellular information, their subcellular location assignments are based on computational prediction. In this work, SignalP4 is used to replace SignalP3 as SignalP4 improves the prediction accuracy ( 21 , 31 ). However, the information generated by SignalP3 was also included as it predicts signal peptide cleavage sites more accurately than SignalP4 ( 21 ). The rules for assigning a protein subcellular location are defined below.
Secreted protein
Secreted proteins are further divided as curated secreted proteins, highly likely secreted, likely secreted, and weakly likely secreted. Curated secreted proteins are proteins that are annotated and reviewed to be ‘secreted’ or ‘extracellular’ in the subcellular location from the UniProtKB/Swiss-Prot dataset. Four predictors consisting of SignalP4, Phobius, TargetP and WoLF PSORT are used for protein secretory signal peptide or subcellular location prediction ( 19 ). The highly likely secreted, likely secreted and weakly likely secreted proteins are proteins that are predicted to be secreted or contain a secretory signal peptide by four and three, two or one of the four tools, respectively. The accuracies for these subcategories of secreted proteins are reported in the section of results. It should be noted that proteins having a transmembrane domain or an ER retention signal were excluded from this set. We recommend that the data for making up a secretome should consist of curated secreted proteins and the predicted highly likely secreted protein dataset. The rational for having subcategories of likely secreted and weakly likely secreted proteins is to provide a means for a user to access these data as some of them may be real secreted proteins.
Mitochondrial proteins
A protein predicted as ‘M’ (for mitochondrial) for subcellular location by TargetP and ‘mito’ by WoLF PSORT is classified as a mitochondrial protein. The accuracy is reported in the result. If it is also classified as a membrane protein by TMHMM, then it is further classified as mitochondrial membrane protein.
ER proteins
ER proteins were predicted using WoLF PSORT and PS-Scan. If they contain one or more transmembrane domains, they are classified as ER membrane proteins. Otherwise, they are classified as ER luminal proteins. Proteins predicted to contain a signal peptide by SignalP 4.0 and an ER target signal (Prosite: PS00014) by PS-Scan often are luminal ER proteins.
GPI-anchored proteins
Signal peptide containing proteins that were predicted to have a GPI anchor by FragAnchor were further classified as GPI-anchored proteins. Protein sequences predicted to have a signal peptide and a GPI anchor may attach to the outer leaflet of the plasma membrane or are secreted, thereby becoming components of the extracellular matrix.
Proteins in other subcellular locations
Other subcellular locations, including cytoplasm (cytosol), cytoskeleton, Golgi apparatus, lysosome, nucleus, peroxisome, plasma membrane and vacuole, were predicted by WoLF PSORT. For a protein predicted as located in Golgi apparatus, nucleus or vacuole, it was further classified as a membrane protein in that specific subcellular location if it contained one or more transmembrane domain predicted by TMHMM.
Database implementation
The protein sequence data, species information, subcellular annotation and information predicted from the tools mentioned above were formatted into tab-delimited text files and were stored in a relational database using MySQL hosted in a Linux server. The user interface and modules to access the data were implemented using PHP. BLAST utility and community annotation submission can be accessed from links on the main user interface at http://proteomics.ysu.edu/secretomes/animal/index.php . The supplementary tables and all other data described in the work can be downloaded at http://proteomics.ysu.edu/publication/data/MetazSecKB/ .
Evaluation of prediction accuracies of protein subcellular locations
Prediction accuracy evaluation of human and animal protein subcellular locations a
| . | TP . | FP . | TN . | FN . | Sn (%) . | Sp (%) . | MCC . |
|---|---|---|---|---|---|---|---|
| (a) Mitochondrial proteins | |||||||
| TargetP | 930 | 972 | 16 032 | 940 | 49.7 | 94.3 | 0.44 |
| WoLF PSORT | 920 | 482 | 16 522 | 950 | 49.2 | 97.2 | 0.53 |
| TargetP AND WoLF PSORT | 794 | 262 | 16 742 | 1076 | 42.5 | 98.5 | 0.53 |
| TaregetP OR WoLF PSORT | 1056 | 1202 | 15 802 | 814 | 56.5 | 92.9 | 0.45 |
| (b) Secreted proteins b | |||||||
| Secreted | 5024 | 276 | 12 874 | 700 | 87.8 | 97.9 | 0.88 |
| S + HLS | 5350 | 522 | 12 628 | 374 | 93.5 | 96.0 | 0.89 |
| S + HLS + LS | 5413 | 794 | 12 356 | 311 | 94.6 | 94.0 | 0.87 |
| S + HLS + LS + WLS | 5440 | 1462 | 11 688 | 284 | 95.0 | 88.9 | 0.80 |
| (c) The subcellular locations | |||||||
| Cytoplasm | 1095 | 1124 | 15 779 | 876 | 55.6 | 93.4 | 0.46 |
| Cytoskeleton | 218 | 63 | 18 020 | 573 | 27.6 | 99.7 | 0.45 |
| ER | 257 | 187 | 17 906 | 524 | 32.9 | 99.0 | 0.42 |
| Golgi | 12 | 21 | 18 584 | 257 | 4.5 | 99.9 | 0.12 |
| Lysosome | 1 | 8 | 18 675 | 190 | 0.5 | 100.0 | 0.02 |
| Nucleus | 2979 | 893 | 14 190 | 812 | 78.6 | 94.1 | 0.72 |
| Peroxisome | 4 | 101 | 18 653 | 116 | 3.3 | 99.5 | 0.03 |
| Plasma membrane | 2767 | 647 | 14 880 | 580 | 82.7 | 95.8 | 0.78 |
| Vacuole | 0 | 0 | 18 855 | 19 | 0.0 | 100.0 | - |
| . | TP . | FP . | TN . | FN . | Sn (%) . | Sp (%) . | MCC . |
|---|---|---|---|---|---|---|---|
| (a) Mitochondrial proteins | |||||||
| TargetP | 930 | 972 | 16 032 | 940 | 49.7 | 94.3 | 0.44 |
| WoLF PSORT | 920 | 482 | 16 522 | 950 | 49.2 | 97.2 | 0.53 |
| TargetP AND WoLF PSORT | 794 | 262 | 16 742 | 1076 | 42.5 | 98.5 | 0.53 |
| TaregetP OR WoLF PSORT | 1056 | 1202 | 15 802 | 814 | 56.5 | 92.9 | 0.45 |
| (b) Secreted proteins b | |||||||
| Secreted | 5024 | 276 | 12 874 | 700 | 87.8 | 97.9 | 0.88 |
| S + HLS | 5350 | 522 | 12 628 | 374 | 93.5 | 96.0 | 0.89 |
| S + HLS + LS | 5413 | 794 | 12 356 | 311 | 94.6 | 94.0 | 0.87 |
| S + HLS + LS + WLS | 5440 | 1462 | 11 688 | 284 | 95.0 | 88.9 | 0.80 |
| (c) The subcellular locations | |||||||
| Cytoplasm | 1095 | 1124 | 15 779 | 876 | 55.6 | 93.4 | 0.46 |
| Cytoskeleton | 218 | 63 | 18 020 | 573 | 27.6 | 99.7 | 0.45 |
| ER | 257 | 187 | 17 906 | 524 | 32.9 | 99.0 | 0.42 |
| Golgi | 12 | 21 | 18 584 | 257 | 4.5 | 99.9 | 0.12 |
| Lysosome | 1 | 8 | 18 675 | 190 | 0.5 | 100.0 | 0.02 |
| Nucleus | 2979 | 893 | 14 190 | 812 | 78.6 | 94.1 | 0.72 |
| Peroxisome | 4 | 101 | 18 653 | 116 | 3.3 | 99.5 | 0.03 |
| Plasma membrane | 2767 | 647 | 14 880 | 580 | 82.7 | 95.8 | 0.78 |
| Vacuole | 0 | 0 | 18 855 | 19 | 0.0 | 100.0 | - |
Note: FP, false positives; FN, false negatives; MCC, Matthews correlation coefficient; Sn, sensitivity; Sp, specificity; TP, true positives; TN, true negatives.
a The dataset contains a total of 18 874 proteins.
b Secreted: predicted by four predictors; HLS: highly likely secreted, predicted by three out of four predictors; LS: likely secreted, predicted by two out of four predictors; WLS: weakly likely secreted, predicted by one out of four predictors.
Prediction accuracy evaluation of human and animal protein subcellular locations a
| . | TP . | FP . | TN . | FN . | Sn (%) . | Sp (%) . | MCC . |
|---|---|---|---|---|---|---|---|
| (a) Mitochondrial proteins | |||||||
| TargetP | 930 | 972 | 16 032 | 940 | 49.7 | 94.3 | 0.44 |
| WoLF PSORT | 920 | 482 | 16 522 | 950 | 49.2 | 97.2 | 0.53 |
| TargetP AND WoLF PSORT | 794 | 262 | 16 742 | 1076 | 42.5 | 98.5 | 0.53 |
| TaregetP OR WoLF PSORT | 1056 | 1202 | 15 802 | 814 | 56.5 | 92.9 | 0.45 |
| (b) Secreted proteins b | |||||||
| Secreted | 5024 | 276 | 12 874 | 700 | 87.8 | 97.9 | 0.88 |
| S + HLS | 5350 | 522 | 12 628 | 374 | 93.5 | 96.0 | 0.89 |
| S + HLS + LS | 5413 | 794 | 12 356 | 311 | 94.6 | 94.0 | 0.87 |
| S + HLS + LS + WLS | 5440 | 1462 | 11 688 | 284 | 95.0 | 88.9 | 0.80 |
| (c) The subcellular locations | |||||||
| Cytoplasm | 1095 | 1124 | 15 779 | 876 | 55.6 | 93.4 | 0.46 |
| Cytoskeleton | 218 | 63 | 18 020 | 573 | 27.6 | 99.7 | 0.45 |
| ER | 257 | 187 | 17 906 | 524 | 32.9 | 99.0 | 0.42 |
| Golgi | 12 | 21 | 18 584 | 257 | 4.5 | 99.9 | 0.12 |
| Lysosome | 1 | 8 | 18 675 | 190 | 0.5 | 100.0 | 0.02 |
| Nucleus | 2979 | 893 | 14 190 | 812 | 78.6 | 94.1 | 0.72 |
| Peroxisome | 4 | 101 | 18 653 | 116 | 3.3 | 99.5 | 0.03 |
| Plasma membrane | 2767 | 647 | 14 880 | 580 | 82.7 | 95.8 | 0.78 |
| Vacuole | 0 | 0 | 18 855 | 19 | 0.0 | 100.0 | - |
| . | TP . | FP . | TN . | FN . | Sn (%) . | Sp (%) . | MCC . |
|---|---|---|---|---|---|---|---|
| (a) Mitochondrial proteins | |||||||
| TargetP | 930 | 972 | 16 032 | 940 | 49.7 | 94.3 | 0.44 |
| WoLF PSORT | 920 | 482 | 16 522 | 950 | 49.2 | 97.2 | 0.53 |
| TargetP AND WoLF PSORT | 794 | 262 | 16 742 | 1076 | 42.5 | 98.5 | 0.53 |
| TaregetP OR WoLF PSORT | 1056 | 1202 | 15 802 | 814 | 56.5 | 92.9 | 0.45 |
| (b) Secreted proteins b | |||||||
| Secreted | 5024 | 276 | 12 874 | 700 | 87.8 | 97.9 | 0.88 |
| S + HLS | 5350 | 522 | 12 628 | 374 | 93.5 | 96.0 | 0.89 |
| S + HLS + LS | 5413 | 794 | 12 356 | 311 | 94.6 | 94.0 | 0.87 |
| S + HLS + LS + WLS | 5440 | 1462 | 11 688 | 284 | 95.0 | 88.9 | 0.80 |
| (c) The subcellular locations | |||||||
| Cytoplasm | 1095 | 1124 | 15 779 | 876 | 55.6 | 93.4 | 0.46 |
| Cytoskeleton | 218 | 63 | 18 020 | 573 | 27.6 | 99.7 | 0.45 |
| ER | 257 | 187 | 17 906 | 524 | 32.9 | 99.0 | 0.42 |
| Golgi | 12 | 21 | 18 584 | 257 | 4.5 | 99.9 | 0.12 |
| Lysosome | 1 | 8 | 18 675 | 190 | 0.5 | 100.0 | 0.02 |
| Nucleus | 2979 | 893 | 14 190 | 812 | 78.6 | 94.1 | 0.72 |
| Peroxisome | 4 | 101 | 18 653 | 116 | 3.3 | 99.5 | 0.03 |
| Plasma membrane | 2767 | 647 | 14 880 | 580 | 82.7 | 95.8 | 0.78 |
| Vacuole | 0 | 0 | 18 855 | 19 | 0.0 | 100.0 | - |
Note: FP, false positives; FN, false negatives; MCC, Matthews correlation coefficient; Sn, sensitivity; Sp, specificity; TP, true positives; TN, true negatives.
a The dataset contains a total of 18 874 proteins.
b Secreted: predicted by four predictors; HLS: highly likely secreted, predicted by three out of four predictors; LS: likely secreted, predicted by two out of four predictors; WLS: weakly likely secreted, predicted by one out of four predictors.
Results
Prediction accuracy evaluation
Mitochondrial proteins
The accuracy results are shown in Table 1 a. When an individual tool was used, WoLF PSORT prediction showed a slightly lower sensitivity but a higher specificity than TargetP prediction. Thus, the MCC value was higher in the set predicted by WoLF PSORT (0.53) than the set predicted by TargetP (0.44). If only positives predicted by both tools were used, the specificity was slightly increased and the MCC value remains unchanged (0.53) compared with WoLF PSORT prediction. In contrast, including positives predicted by either tool decreased the MCC value to 0.45. Thus we assigned mitochondrial subcellular locations to entries only predicted to be mitochondrial proteins by both programs. As the specificity was high (up to 98.5%) when both tools were used, these predicted entries were reasonably reliable. However, the prediction sensitivity (42.5%) of the tools was low, i.e. more than half of proteins located in mitochondria remained to be predicted. Thus future efforts need to be made to improve prediction sensitivity for mitochondrial proteins.
Secreted proteins
Our previous evaluation showed that secreted prediction accuracy can be improved by removing transmembrane proteins, which can be predicted using TMHMM, and ER resident proteins, which can be predicted using PS-Scan ( 19 ). As we employed four tools—SignalP (version 4), TargetP, WoLF PSORT and Phobius—for predicting secreted proteins or secretory signal peptides, we had to determine which should be included in the secretome set. After removing transmembrane proteins and ER proteins, the protein set predicted either to contain a secretory signal peptide or to be secreted are divided into four categories: (i) Secreted: predicted by 4 predictors; (ii) Highly likely secreted (HLS): predicted by 3 out of 4 predictors; (iii) Likely secreted (LS): predicted by 2 out of 4 predictors; and (iv) Weakly likely secreted (WLS): predicted by 1 out of 4 predictors. The dataset consisted of 5724 curated secreted proteins as positives and 13 150 proteins located in other subcellular locations as negatives. The accuracy results are shown in Table 1 b.
As expected, when only entries were predicted by all four tools to be positives as true positives, the prediction specificity was increased. However, the sensitivity was decreased. On the other hand, the prediction specificity was decreased but the sensitivity was increased when including all entries predicted by any of the four tools to be positives as true positives. Based on the MCC values, the most accurate prediction (0.89) for a secretome includes secreted entries predicted by at least three out of four predictors with a specificity of 96.0% and a sensitivity of 93.5% ( Table 1 b). Thus, we recommend including only curated secreted proteins and highly likely secreted proteins for estimating the secretome size. Though including the set of likely secreted proteins increased the coverage of a secretome, it increased more (272 entries) false positives than true (63 entries) positives. It should be noted that both entries predicted by 4 of 4 tools and 3 of 4 tools were assigned as the category of highly like secreted in the database, making them distinguishable from curated secreted entries.
Proteins in other subcellular locations
Proteins for the cytoplasm subset also include cytosol as these two terms are used interchangeably in the UniProtKB annotation. However, we noticed that the annotated cytoskeleton entries are also annotated as cytoplasm. In our evaluation, cytoskeleton proteins were not counted in the subset of cytoplasm. We would also like to point out that plasma membrane proteins were annotated as cell membrane in UniProtKB, thus cell membrane proteins were retrieved for evaluating the category of plasma membrane. The prediction accuracy results for proteins located in cytoplasm, cytoskeleton, ER, Golgi apparatus, lysosome, nucleus, peroxisome, plasma membrane and vacuole are shown in Table 1 c.
The prediction accuracies for these subcellular locations vary significantly. Predictions of proteins located in nucleus and plasma membrane were relatively accurate with a MCC value of 0.78 and 0.72, respectively. Predictions for proteins located in cytoplasm, cytoskeleton, and ER were highly specific (specificity 93.4–99.7%) with a MCC value of 0.42–0.46. However, the sensitivities (27.6–55.6%) need to be improved for these subcellular locations. Predictions for proteins located in Golgi apparatus, lysosome, peroxisome were also highly specific (specificity > 99%) but with a very low sensitivity (0.5–4.5%). Human and animal vacuolar proteins could not be predicted by WoLF PSORT as there were no positive being predicted ( Table 1 c). It should be noted that the low MCC values for some of the subcellular locations were caused by low sensitivities, and in fact, the specificities were relatively high. Thus, there are a good number of proteins located in these subcellular locations not being predicted. However, if a protein is predicted to be located in such a location, the prediction is most likely reliable.
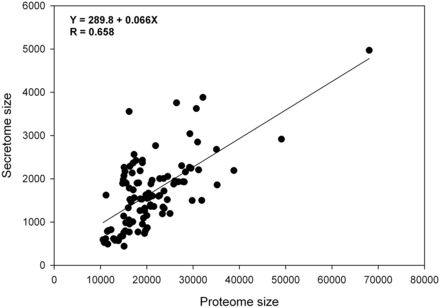
Database statistics: subcellular proteome distribution in different species
The database contains curated and predicted subcellular location information of 4 080 818 metazoan proteins that were downloaded from UniProtKB. These proteins were generated from 185 256 metazoa species and subspecies with 121 of them having a complete proteome. Species specific proteins located at each subcellular location can be searched and downloaded from the database user interface. The distributions of subcellular proteomes in human and different animal species having a complete proteome are summarized in Table 2 and Supplementary Table S1 . Table 2 includes the following subcellular locations: secreted proteins (3 subcategories), mitochondrial membrane and mitochondrial non-membrane, cytoplasm (cytosol), nuclear membrane and nuclear non-membrane, plasma membrane. The category of secreted proteins includes the following subcategories: curated secreted, highly likely secreted and likely secreted. Information on other subcellular protein locations including weakly likely secreted, cytoskeleton, ER (membrane or lumen), Golgi apparatus (membrane or lumen), lysosome, peroxisome, vacuole (membrane or non-membrane), other membrane, other curated locations and the information of species taxonomy can be found in Supplementary Table S1 .
Summary of proteins located in some major subcellular locations in human and different animal species
| . | Referenceproteome . | Totalproteins . | Curatedsecreted . | Predicted . | Mito . | . | Nuc . | Plasmamem . | . | Secr(%) . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | HLS . | LS . | mem . | non-mem . | Cyt . | mem . | non-mem . | . | Secr . | . | |
| Vertebrata (Actinopterygii) | |||||||||||||
| Oryzias latipes | 24 633 | 26 060 | 144 | 1805 | 649 | 141 | 1185 | 7330 | 162 | 8629 | 3580 | 1949 | 7.5 |
| Xiphophorus maculatus | 20 451 | 20 527 | 92 | 1476 | 510 | 73 | 959 | 5288 | 92 | 7237 | 3052 | 1568 | 7.6 |
| Oreochromis niloticus | 26 753 | 27 551 | 122 | 2179 | 638 | 148 | 1051 | 6971 | 149 | 9638 | 4078 | 2301 | 8.4 |
| Gasterosteus aculeatus | 27 248 | 28 110 | 114 | 1813 | 618 | 106 | 1418 | 8080 | 142 | 9443 | 3877 | 1927 | 6.9 |
| Takifugu rubripes | 47 856 | 49 090 | 261 | 2655 | 1028 | 172 | 1645 | 12 630 | 323 | 17 843 | 8443 | 2916 | 5.9 |
| Tetraodon nigroviridis* | 23 073 | 49 327 | 194 | 2700 | 1236 | 182 | 2248 | 13 333 | 337 | 16 961 | 5944 | 2894 | 5.9 |
| Danio rerio* | 41 054 | 55 414 | 372 | 4635 | 1189 | 282 | 2319 | 14 909 | 267 | 19 521 | 6866 | 5007 | 9.0 |
| Vertebrata (Amphibia) | |||||||||||||
| Xenopus tropicalis* | 23 491 | 30 521 | 194 | 1926 | 656 | 169 | 1327 | 9674 | 168 | 10 086 | 4113 | 2120 | 6.9 |
| X. laevis | 16 011 | 269 | 1059 | 262 | 161 | 752 | 5124 | 109 | 5711 | 1636 | 1328 | 8.3 | |
| Vertebrata (Mammalia) | |||||||||||||
| Glires | |||||||||||||
| Oryctolagus cuniculus | 21 150 | 22 788 | 334 | 1670 | 479 | 135 | 1222 | 5692 | 150 | 7241 | 3665 | 2004 | 8.8 |
| Heterocephalus glaber | 21 449 | 21 548 | 93 | 1266 | 513 | 90 | 1009 | 6343 | 103 | 6924 | 3266 | 1359 | 6.3 |
| Cavia porcellus | 19 911 | 20 378 | 236 | 1432 | 461 | 100 | 1016 | 5349 | 103 | 6410 | 3342 | 1668 | 8.2 |
| Cricetulus griseus | 23 884 | 24 442 | 109 | 1407 | 927 | 96 | 1170 | 7073 | 116 | 7114 | 2786 | 1516 | 6.2 |
| Mus musculus* | 43 539 | 74 158 | 1792 | 4350 | 1698 | 717 | 3443 | 20 456 | 714 | 23 226 | 9137 | 6142 | 8.3 |
| Rattus norvegicus* | 27 340 | 33 555 | 966 | 2211 | 637 | 476 | 1473 | 9153 | 411 | 10 094 | 5407 | 3177 | 9.5 |
| Spermophilus tridecemlineatus | 19 966 | 20 079 | 110 | 1437 | 429 | 83 | 937 | 5488 | 103 | 6603 | 3290 | 1547 | 7.7 |
| Primates | |||||||||||||
| Macaca fascicularis * | 17 396 | 28 955 | 233 | 1912 | 928 | 359 | 1976 | 7639 | 121 | 8186 | 2970 | 2145 | 7.4 |
| M. mulatta* | 35 536 | 69 567 | 407 | 4554 | 1694 | 653 | 3719 | 18 667 | 326 | 23 502 | 7295 | 4961 | 7.1 |
| Gorilla gorilla gorilla | 27 286 | 27 371 | 212 | 1994 | 676 | 218 | 1480 | 6701 | 151 | 9481 | 3358 | 2206 | 8.1 |
| Homo sapiens* | 68 049 | 13 5661 | 2020 | 6682 | 3480 | 3737 | 17 623 | 34 825 | 877 | 34 274 | 10 607 | 8702 | 6.4 |
| Pan troglodytes* | 20 126 | 33 326 | 296 | 2241 | 820 | 447 | 1825 | 7966 | 137 | 11 618 | 4190 | 2537 | 7.6 |
| Pongo abelii | 22 785 | 24 529 | 237 | 1818 | 580 | 229 | 1457 | 6452 | 168 | 8228 | 2879 | 2055 | 8.4 |
| Nomascus leucogenys | 19 734 | 19 837 | 141 | 1489 | 518 | 114 | 1143 | 5053 | 99 | 6893 | 2457 | 1630 | 8.2 |
| Callithrix jacchus* | 42 025 | 55 085 | 244 | 3776 | 1280 | 195 | 2867 | 15 064 | 308 | 20 159 | 6178 | 4020 | 7.3 |
| Otolemur garnettii | 19 930 | 20 156 | 99 | 1515 | 480 | 93 | 1022 | 5226 | 96 | 6801 | 3099 | 1614 | 8.0 |
| Carnivora | |||||||||||||
| Canis familiaris | 25 439 | 28 362 | 345 | 1813 | 595 | 385 | 1491 | 7040 | 170 | 9489 | 4047 | 2158 | 7.6 |
| Mustela putorius furo | 38 826 | 173 | 2017 | 984 | 137 | 2127 | 11 785 | 154 | 12 830 | 4073 | 2190 | 5.6 | |
| Neovison vison | 16 237 | 18 | 750 | 356 | 66 | 839 | 5636 | 52 | 5233 | 1507 | 768 | 4.7 | |
| Ailuropoda melanoleuca* | 21 136 | 35 743 | 247 | 2086 | 779 | 176 | 1746 | 9975 | 162 | 11 905 | 5133 | 2333 | 6.5 |
| Felis catus | 20 303 | 21 230 | 196 | 1406 | 483 | 108 | 1065 | 5831 | 107 | 6791 | 3091 | 1602 | 7.5 |
| Cetartiodactyla | |||||||||||||
| Bos mutus | 18 922 | 150 | 1377 | 453 | 123 | 931 | 4911 | 85 | 5854 | 3159 | 1527 | 8.1 | |
| Bos taurus* | 23 842 | 31 780 | 880 | 2215 | 620 | 508 | 1491 | 8171 | 317 | 9598 | 4492 | 3095 | 9.7 |
| Sus scrofa* | 26 054 | 33 962 | 645 | 2534 | 779 | 411 | 1560 | 9038 | 166 | 9463 | 4787 | 3179 | 9.4 |
| Camelus ferus | 20 028 | 67 | 1084 | 636 | 99 | 1132 | 5715 | 147 | 6257 | 2588 | 1151 | 5.7 | |
| Chiroptera | |||||||||||||
| Pteropus alecto | 19 520 | 19 548 | 97 | 1162 | 488 | 74 | 1160 | 5364 | 121 | 6774 | 2447 | 1259 | 6.4 |
| Myotis brandtii | 19 301 | 58 | 1032 | 432 | 90 | 938 | 5806 | 104 | 6427 | 2250 | 1090 | 5.6 | |
| M. davidii | 15 446 | 15 466 | 67 | 916 | 345 | 60 | 782 | 4530 | 73 | 5194 | 1816 | 983 | 6.4 |
| M. lucifugus | 20 650 | 20 899 | 143 | 1738 | 431 | 100 | 1052 | 5716 | 96 | 6782 | 2855 | 1881 | 9.0 |
| Other mammalia | |||||||||||||
| Loxodonta africana | 25 615 | 25 832 | 132 | 1744 | 556 | 128 | 1119 | 6554 | 129 | 8459 | 4835 | 1876 | 7.3 |
| Equus caballus | 22 676 | 27 841 | 272 | 1659 | 514 | 284 | 1042 | 8886 | 133 | 8701 | 3825 | 1931 | 6.9 |
| Tupaia chinensis | 20 824 | 20 851 | 85 | 1275 | 527 | 64 | 1149 | 5699 | 125 | 6701 | 3114 | 1360 | 6.5 |
| Sarcophilus harrisii | 22 388 | 22 565 | 107 | 1490 | 553 | 110 | 867 | 6368 | 102 | 7495 | 3495 | 1597 | 7.1 |
| Monodelphis domestica | 22 240 | 22 794 | 108 | 1505 | 485 | 84 | 1103 | 6398 | 106 | 7252 | 3930 | 1613 | 7.1 |
| Ornithorhynchus anatinus | 23 552 | 23 763 | 113 | 1202 | 698 | 103 | 1111 | 7229 | 95 | 7184 | 3157 | 1315 | 5.5 |
| Vertebrata (Testudines + Archosauria group) | |||||||||||||
| Anas platyrhynchos * | 16 377 | 31 879 | 139 | 1360 | 893 | 123 | 1269 | 10 542 | 148 | 9829 | 3316 | 1499 | 4.7 |
| Meleagris gallopavo | 16 537 | 16 991 | 114 | 892 | 413 | 75 | 673 | 5622 | 83 | 5377 | 2073 | 1006 | 5.9 |
| Gallus gallus* | 17 623 | 23 800 | 440 | 1640 | 581 | 278 | 1231 | 6403 | 147 | 7077 | 3282 | 2080 | 8.7 |
| Ficedula albicollis | 15 922 | 16 148 | 64 | 985 | 390 | 57 | 778 | 4669 | 81 | 5208 | 2021 | 1049 | 6.5 |
| Taeniopygia guttata | 18 141 | 19 724 | 85 | 716 | 432 | 77 | 972 | 6749 | 72 | 6197 | 2211 | 801 | 4.1 |
| Chelonia mydas | 19 066 | 71 | 880 | 478 | 71 | 794 | 6031 | 97 | 6384 | 2194 | 951 | 5.0 | |
| Pelodiscus sinensis | 20 784 | 126 | 1271 | 492 | 64 | 798 | 6724 | 92 | 6703 | 2683 | 1397 | 6.7 | |
| Other vertebrata | |||||||||||||
| Petromyzon marinus | 13 160 | 54 | 522 | 255 | 66 | 669 | 3945 | 34 | 3797 | 1239 | 576 | 4.4 | |
| Latimeria chalumnae | 23 429 | 23 513 | 75 | 1270 | 593 | 87 | 993 | 8117 | 116 | 7349 | 2905 | 1345 | 5.7 |
| Anolis carolinensis | 19 109 | 19 562 | 81 | 1238 | 510 | 288 | 936 | 5727 | 89 | 6435 | 2478 | 1319 | 6.7 |
| Invertebrate | |||||||||||||
| Chordata (Tunicata) | |||||||||||||
| Oikopleura dioica* | 17 050 | 29 057 | 15 | 1864 | 1177 | 116 | 1493 | 10 785 | 92 | 8060 | 2540 | 1879 | 6.5 |
| Ciona intestinalis | 17 308 | 18 639 | 28 | 1507 | 601 | 95 | 738 | 6231 | 47 | 5014 | 1812 | 1535 | 8.2 |
| C. savignyi | 20 004 | 20 117 | 45 | 822 | 359 | 52 | 697 | 7792 | 55 | 5685 | 2535 | 867 | 4.3 |
| Ecdysozoa (Arachnida) | |||||||||||||
| Tetranychus urticae | 18 082 | 18 243 | 12 | 1891 | 685 | 95 | 701 | 5986 | 59 | 3615 | 2219 | 1903 | 10.4 |
| Ixodes ricinus | 16 199 | 1 | 3554 | 819 | 136 | 848 | 3528 | 97 | 3649 | 1246 | 3555 | 21.9 | |
| I. scapularis | 20 473 | 21 162 | 21 | 1943 | 762 | 103 | 1433 | 5706 | 95 | 5905 | 1681 | 1964 | 9.3 |
| Rhipicephalus pulchellus | 11 205 | 1 | 1620 | 459 | 60 | 962 | 2380 | 49 | 3424 | 1231 | 1621 | 14.5 | |
| Ecdysozoa (Insecta) | |||||||||||||
| Drosophila mojavensis | 14 525 | 15 086 | 21 | 2049 | 347 | 95 | 832 | 4302 | 74 | 4241 | 1700 | 2070 | 13.7 |
| D. virilis | 14 456 | 14 928 | 34 | 1941 | 354 | 84 | 770 | 4308 | 57 | 4323 | 1709 | 1975 | 13.2 |
| D. erecta | 15 116 | 44 | 2220 | 362 | 71 | 873 | 3918 | 59 | 4366 | 1721 | 2264 | 15.0 | |
| D. grimshawi | 14 754 | 14 798 | 31 | 1861 | 362 | 62 | 773 | 4336 | 79 | 4195 | 1698 | 1892 | 12.8 |
| D. ananassae | 14 968 | 15 298 | 28 | 2139 | 349 | 64 | 791 | 4243 | 67 | 4503 | 1793 | 2167 | 14.2 |
| D. melanogaster* | 20 120 | 39 951 | 254 | 4761 | 923 | 269 | 2127 | 10 613 | 191 | 12 101 | 4659 | 5015 | 12.6 |
| D. persimilis | 16 754 | 16 861 | 28 | 2106 | 420 | 77 | 930 | 4846 | 68 | 5076 | 1701 | 2134 | 12.7 |
| D. pseudoobscura pseudoobscura | 17 047 | 48 | 2316 | 416 | 77 | 939 | 4632 | 71 | 5126 | 1950 | 2364 | 13.9 | |
| D. sechellia | 16 134 | 16 361 | 37 | 2250 | 410 | 71 | 936 | 4464 | 54 | 4765 | 1734 | 2287 | 14.0 |
| D. simulans | 15 354 | 19 057 | 57 | 2372 | 436 | 100 | 1028 | 5483 | 56 | 5374 | 2165 | 2429 | 12.7 |
| D. willistoni | 15 447 | 15 564 | 25 | 1875 | 355 | 77 | 815 | 4808 | 60 | 4434 | 1722 | 1900 | 12.2 |
| D. yakuba | 17 257 | 41 | 2521 | 392 | 77 | 1006 | 4752 | 59 | 5024 | 1798 | 2562 | 14.8 | |
| Megaselia scalaris | 11 463 | 11 503 | 10 | 773 | 417 | 31 | 449 | 4947 | 24 | 2402 | 639 | 783 | 6.8 |
| Anopheles darlingi | 10 447 | 11 686 | 8 | 793 | 272 | 93 | 758 | 3687 | 54 | 3971 | 1162 | 801 | 6.9 |
| A. gambiae* | 13 072 | 19 384 | 50 | 2610 | 410 | 87 | 1104 | 6027 | 57 | 4834 | 1869 | 2660 | 13.7 |
| Aedes aegypti | 16 654 | 17 683 | 54 | 2367 | 469 | 182 | 961 | 5052 | 56 | 4873 | 2008 | 2421 | 13.7 |
| Culex quinquefasciatus | 18 703 | 19 062 | 25 | 2345 | 534 | 128 | 1104 | 5751 | 73 | 5501 | 1866 | 2370 | 12.4 |
| Dendroctonus ponderosae | 23 650 | 14 | 1992 | 549 | 106 | 1153 | 8928 | 96 | 6087 | 2502 | 2006 | 8.5 | |
| Tribolium castaneum | 16 502 | 17 074 | 26 | 1717 | 423 | 66 | 846 | 5830 | 50 | 4196 | 2109 | 1743 | 10.2 |
| Apis mellifera | 10 910 | 12 299 | 65 | 757 | 267 | 81 | 390 | 4440 | 45 | 3293 | 1714 | 822 | 6.7 |
| Camponotus floridanus | 14 787 | 14 801 | 15 | 662 | 329 | 45 | 744 | 5533 | 65 | 4078 | 1346 | 677 | 4.6 |
| Acromyrmex echinatior | 13 962 | 13 970 | 17 | 592 | 327 | 36 | 847 | 5226 | 62 | 4219 | 1253 | 609 | 4.4 |
| Atta cephalotes | 18 079 | 18 113 | 16 | 753 | 597 | 99 | 1094 | 6579 | 75 | 4715 | 1559 | 769 | 4.2 |
| Solenopsis invicta | 14 193 | 14 359 | 26 | 636 | 437 | 100 | 748 | 5413 | 31 | 3508 | 1120 | 662 | 4.6 |
| Harpegnathos saltator | 15 029 | 15 042 | 17 | 739 | 329 | 46 | 696 | 5484 | 60 | 4223 | 1299 | 756 | 5.0 |
| Nasonia vitripennis | 17 040 | 17 289 | 14 | 1545 | 305 | 65 | 701 | 6951 | 55 | 4883 | 1423 | 1559 | 9.0 |
| Bombyx mori | 14 767 | 17 915 | 125 | 1773 | 379 | 108 | 806 | 6293 | 54 | 4580 | 1681 | 1898 | 10.6 |
| Danaus plexippus | 16 253 | 16 358 | 34 | 1486 | 441 | 95 | 808 | 5657 | 66 | 4528 | 1493 | 1520 | 9.3 |
| Pararge aegeria | 15 104 | 12 | 428 | 561 | 75 | 850 | 5983 | 14 | 3763 | 503 | 440 | 2.9 | |
| Rhodnius prolixus | 15 180 | 16 639 | 44 | 1420 | 537 | 41 | 562 | 6782 | 62 | 3769 | 1473 | 1464 | 8.8 |
| Acyrthosiphon pisum | 35 809 | 35 211 | 24 | 1834 | 814 | 102 | 1736 | 15 209 | 66 | 8622 | 1736 | 1858 | 5.3 |
| Pediculus humanus subsp. corporis | 10 847 | 11 | 513 | 257 | 37 | 349 | 4294 | 40 | 3174 | 1193 | 524 | 4.8 | |
| Ecdysozoa (Nematoda) | |||||||||||||
| Ascaris suum* | 9213 | 18 539 | 39 | 1223 | 577 | 107 | 1302 | 5894 | 75 | 4965 | 2437 | 1262 | 6.8 |
| Pristionchus pacificus | 29 079 | 29 319 | 14 | 3027 | 1038 | 75 | 1368 | 9699 | 94 | 7157 | 3263 | 3041 | 10.4 |
| Caenorhabditis brenneri | 29 982 | 30 712 | 21 | 3602 | 896 | 76 | 1134 | 10 314 | 88 | 7338 | 4255 | 3623 | 11.8 |
| C. briggsae | 21 751 | 21 914 | 30 | 2734 | 655 | 119 | 874 | 6435 | 106 | 5178 | 3540 | 2764 | 12.6 |
| C. elegans | 26 173 | 26 447 | 182 | 3573 | 856 | 163 | 1065 | 7156 | 107 | 6190 | 4401 | 3755 | 14.2 |
| C. japonica | 35 063 | 35 069 | 14 | 2665 | 998 | 95 | 2061 | 12 267 | 123 | 9112 | 3234 | 2679 | 7.6 |
| C. remanei | 31 252 | 32 133 | 21 | 3859 | 1117 | 84 | 1282 | 10 352 | 93 | 7199 | 4981 | 3880 | 12.1 |
| Haemonchus contortus | 18 580 | 3 | 2181 | 558 | 83 | 1016 | 5826 | 77 | 4679 | 2261 | 2184 | 11.8 | |
| Brugia malayi* | 1643 | 11 561 | 10 | 668 | 347 | 37 | 579 | 4540 | 44 | 3139 | 908 | 678 | 5.9 |
| Loa loa | 15 319 | 15 356 | 11 | 784 | 588 | 46 | 750 | 5774 | 49 | 3749 | 1387 | 795 | 5.2 |
| Wuchereria bancrofti | 19 298 | 19 525 | 18 | 716 | 677 | 129 | 870 | 8254 | 39 | 4504 | 1205 | 734 | 3.8 |
| Trichinella spiralis | 16 041 | 16 278 | 17 | 935 | 770 | 73 | 980 | 5389 | 71 | 3433 | 1234 | 952 | 5.8 |
| Ecdysozoa (Arthropoda) | |||||||||||||
| Daphnia pulex | 30 137 | 30 988 | 22 | 2827 | 892 | 333 | 1432 | 11 433 | 88 | 8315 | 2130 | 2849 | 9.2 |
| Strigamia maritima | 14 972 | 15 011 | 19 | 1118 | 428 | 48 | 726 | 5331 | 68 | 3635 | 1835 | 1137 | 7.6 |
| Lophotrochozoa | |||||||||||||
| Helobdella robusta | 23 328 | 23 379 | 19 | 1170 | 671 | 59 | 924 | 9385 | 145 | 5866 | 2226 | 1189 | 5.1 |
| Capitella teleta | 31 207 | 22 | 2183 | 907 | 76 | 1263 | 10 827 | 106 | 7765 | 3917 | 2205 | 7.1 | |
| Crassostrea gigas | 25 982 | 26 850 | 26 | 1904 | 633 | 85 | 814 | 10 178 | 140 | 7045 | 2912 | 1930 | 7.2 |
| Lottia gigantea | 23 721 | 34 | 1683 | 588 | 48 | 659 | 9382 | 76 | 5530 | 2734 | 1717 | 7.2 | |
| Platyhelminthes | |||||||||||||
| Echinococcus granulosus | 11 124 | 0 | 614 | 375 | 381 | 656 | 2855 | 40 | 3518 | 1260 | 614 | 5.5 | |
| E. multilocularis | 10 572 | 0 | 591 | 326 | 91 | 656 | 2878 | 48 | 3532 | 1239 | 591 | 5.6 | |
| Clonorchis sinensis | 13 606 | 13 880 | 6 | 562 | 349 | 55 | 990 | 4294 | 89 | 5074 | 1234 | 568 | 4.1 |
| Schistosoma japonicum | 16 236 | 17 | 1767 | 607 | 70 | 853 | 6086 | 36 | 3511 | 1357 | 1784 | 11.0 | |
| S. mansoni | 11 723 | 12 836 | 9 | 605 | 427 | 203 | 505 | 4491 | 60 | 3740 | 1242 | 614 | 4.8 |
| Other Invertebrates | |||||||||||||
| Amphimedon queenslandica | 29 741 | 29 816 | 6 | 1490 | 893 | 65 | 1246 | 11 722 | 73 | 7333 | 2685 | 1496 | 5.0 |
| Nematostella vectensis | 24 435 | 25 035 | 61 | 1135 | 586 | 58 | 1005 | 8651 | 72 | 6385 | 3293 | 1196 | 4.8 |
| Strongylocentrotus purpuratus | 28 567 | 29 560 | 46 | 2198 | 737 | 94 | 1101 | 9895 | 145 | 8580 | 4026 | 2244 | 7.6 |
| Trichoplax adhaerens | 11 520 | 11 590 | 7 | 482 | 213 | 36 | 489 | 5013 | 44 | 2502 | 1776 | 489 | 4.2 |
| Branchiostoma floridae | 28 544 | 29 237 | 37 | 2227 | 710 | 152 | 1146 | 8799 | 140 | 7800 | 3826 | 2264 | 7.7 |
| . | Referenceproteome . | Totalproteins . | Curatedsecreted . | Predicted . | Mito . | . | Nuc . | Plasmamem . | . | Secr(%) . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | HLS . | LS . | mem . | non-mem . | Cyt . | mem . | non-mem . | . | Secr . | . | |
| Vertebrata (Actinopterygii) | |||||||||||||
| Oryzias latipes | 24 633 | 26 060 | 144 | 1805 | 649 | 141 | 1185 | 7330 | 162 | 8629 | 3580 | 1949 | 7.5 |
| Xiphophorus maculatus | 20 451 | 20 527 | 92 | 1476 | 510 | 73 | 959 | 5288 | 92 | 7237 | 3052 | 1568 | 7.6 |
| Oreochromis niloticus | 26 753 | 27 551 | 122 | 2179 | 638 | 148 | 1051 | 6971 | 149 | 9638 | 4078 | 2301 | 8.4 |
| Gasterosteus aculeatus | 27 248 | 28 110 | 114 | 1813 | 618 | 106 | 1418 | 8080 | 142 | 9443 | 3877 | 1927 | 6.9 |
| Takifugu rubripes | 47 856 | 49 090 | 261 | 2655 | 1028 | 172 | 1645 | 12 630 | 323 | 17 843 | 8443 | 2916 | 5.9 |
| Tetraodon nigroviridis* | 23 073 | 49 327 | 194 | 2700 | 1236 | 182 | 2248 | 13 333 | 337 | 16 961 | 5944 | 2894 | 5.9 |
| Danio rerio* | 41 054 | 55 414 | 372 | 4635 | 1189 | 282 | 2319 | 14 909 | 267 | 19 521 | 6866 | 5007 | 9.0 |
| Vertebrata (Amphibia) | |||||||||||||
| Xenopus tropicalis* | 23 491 | 30 521 | 194 | 1926 | 656 | 169 | 1327 | 9674 | 168 | 10 086 | 4113 | 2120 | 6.9 |
| X. laevis | 16 011 | 269 | 1059 | 262 | 161 | 752 | 5124 | 109 | 5711 | 1636 | 1328 | 8.3 | |
| Vertebrata (Mammalia) | |||||||||||||
| Glires | |||||||||||||
| Oryctolagus cuniculus | 21 150 | 22 788 | 334 | 1670 | 479 | 135 | 1222 | 5692 | 150 | 7241 | 3665 | 2004 | 8.8 |
| Heterocephalus glaber | 21 449 | 21 548 | 93 | 1266 | 513 | 90 | 1009 | 6343 | 103 | 6924 | 3266 | 1359 | 6.3 |
| Cavia porcellus | 19 911 | 20 378 | 236 | 1432 | 461 | 100 | 1016 | 5349 | 103 | 6410 | 3342 | 1668 | 8.2 |
| Cricetulus griseus | 23 884 | 24 442 | 109 | 1407 | 927 | 96 | 1170 | 7073 | 116 | 7114 | 2786 | 1516 | 6.2 |
| Mus musculus* | 43 539 | 74 158 | 1792 | 4350 | 1698 | 717 | 3443 | 20 456 | 714 | 23 226 | 9137 | 6142 | 8.3 |
| Rattus norvegicus* | 27 340 | 33 555 | 966 | 2211 | 637 | 476 | 1473 | 9153 | 411 | 10 094 | 5407 | 3177 | 9.5 |
| Spermophilus tridecemlineatus | 19 966 | 20 079 | 110 | 1437 | 429 | 83 | 937 | 5488 | 103 | 6603 | 3290 | 1547 | 7.7 |
| Primates | |||||||||||||
| Macaca fascicularis * | 17 396 | 28 955 | 233 | 1912 | 928 | 359 | 1976 | 7639 | 121 | 8186 | 2970 | 2145 | 7.4 |
| M. mulatta* | 35 536 | 69 567 | 407 | 4554 | 1694 | 653 | 3719 | 18 667 | 326 | 23 502 | 7295 | 4961 | 7.1 |
| Gorilla gorilla gorilla | 27 286 | 27 371 | 212 | 1994 | 676 | 218 | 1480 | 6701 | 151 | 9481 | 3358 | 2206 | 8.1 |
| Homo sapiens* | 68 049 | 13 5661 | 2020 | 6682 | 3480 | 3737 | 17 623 | 34 825 | 877 | 34 274 | 10 607 | 8702 | 6.4 |
| Pan troglodytes* | 20 126 | 33 326 | 296 | 2241 | 820 | 447 | 1825 | 7966 | 137 | 11 618 | 4190 | 2537 | 7.6 |
| Pongo abelii | 22 785 | 24 529 | 237 | 1818 | 580 | 229 | 1457 | 6452 | 168 | 8228 | 2879 | 2055 | 8.4 |
| Nomascus leucogenys | 19 734 | 19 837 | 141 | 1489 | 518 | 114 | 1143 | 5053 | 99 | 6893 | 2457 | 1630 | 8.2 |
| Callithrix jacchus* | 42 025 | 55 085 | 244 | 3776 | 1280 | 195 | 2867 | 15 064 | 308 | 20 159 | 6178 | 4020 | 7.3 |
| Otolemur garnettii | 19 930 | 20 156 | 99 | 1515 | 480 | 93 | 1022 | 5226 | 96 | 6801 | 3099 | 1614 | 8.0 |
| Carnivora | |||||||||||||
| Canis familiaris | 25 439 | 28 362 | 345 | 1813 | 595 | 385 | 1491 | 7040 | 170 | 9489 | 4047 | 2158 | 7.6 |
| Mustela putorius furo | 38 826 | 173 | 2017 | 984 | 137 | 2127 | 11 785 | 154 | 12 830 | 4073 | 2190 | 5.6 | |
| Neovison vison | 16 237 | 18 | 750 | 356 | 66 | 839 | 5636 | 52 | 5233 | 1507 | 768 | 4.7 | |
| Ailuropoda melanoleuca* | 21 136 | 35 743 | 247 | 2086 | 779 | 176 | 1746 | 9975 | 162 | 11 905 | 5133 | 2333 | 6.5 |
| Felis catus | 20 303 | 21 230 | 196 | 1406 | 483 | 108 | 1065 | 5831 | 107 | 6791 | 3091 | 1602 | 7.5 |
| Cetartiodactyla | |||||||||||||
| Bos mutus | 18 922 | 150 | 1377 | 453 | 123 | 931 | 4911 | 85 | 5854 | 3159 | 1527 | 8.1 | |
| Bos taurus* | 23 842 | 31 780 | 880 | 2215 | 620 | 508 | 1491 | 8171 | 317 | 9598 | 4492 | 3095 | 9.7 |
| Sus scrofa* | 26 054 | 33 962 | 645 | 2534 | 779 | 411 | 1560 | 9038 | 166 | 9463 | 4787 | 3179 | 9.4 |
| Camelus ferus | 20 028 | 67 | 1084 | 636 | 99 | 1132 | 5715 | 147 | 6257 | 2588 | 1151 | 5.7 | |
| Chiroptera | |||||||||||||
| Pteropus alecto | 19 520 | 19 548 | 97 | 1162 | 488 | 74 | 1160 | 5364 | 121 | 6774 | 2447 | 1259 | 6.4 |
| Myotis brandtii | 19 301 | 58 | 1032 | 432 | 90 | 938 | 5806 | 104 | 6427 | 2250 | 1090 | 5.6 | |
| M. davidii | 15 446 | 15 466 | 67 | 916 | 345 | 60 | 782 | 4530 | 73 | 5194 | 1816 | 983 | 6.4 |
| M. lucifugus | 20 650 | 20 899 | 143 | 1738 | 431 | 100 | 1052 | 5716 | 96 | 6782 | 2855 | 1881 | 9.0 |
| Other mammalia | |||||||||||||
| Loxodonta africana | 25 615 | 25 832 | 132 | 1744 | 556 | 128 | 1119 | 6554 | 129 | 8459 | 4835 | 1876 | 7.3 |
| Equus caballus | 22 676 | 27 841 | 272 | 1659 | 514 | 284 | 1042 | 8886 | 133 | 8701 | 3825 | 1931 | 6.9 |
| Tupaia chinensis | 20 824 | 20 851 | 85 | 1275 | 527 | 64 | 1149 | 5699 | 125 | 6701 | 3114 | 1360 | 6.5 |
| Sarcophilus harrisii | 22 388 | 22 565 | 107 | 1490 | 553 | 110 | 867 | 6368 | 102 | 7495 | 3495 | 1597 | 7.1 |
| Monodelphis domestica | 22 240 | 22 794 | 108 | 1505 | 485 | 84 | 1103 | 6398 | 106 | 7252 | 3930 | 1613 | 7.1 |
| Ornithorhynchus anatinus | 23 552 | 23 763 | 113 | 1202 | 698 | 103 | 1111 | 7229 | 95 | 7184 | 3157 | 1315 | 5.5 |
| Vertebrata (Testudines + Archosauria group) | |||||||||||||
| Anas platyrhynchos * | 16 377 | 31 879 | 139 | 1360 | 893 | 123 | 1269 | 10 542 | 148 | 9829 | 3316 | 1499 | 4.7 |
| Meleagris gallopavo | 16 537 | 16 991 | 114 | 892 | 413 | 75 | 673 | 5622 | 83 | 5377 | 2073 | 1006 | 5.9 |
| Gallus gallus* | 17 623 | 23 800 | 440 | 1640 | 581 | 278 | 1231 | 6403 | 147 | 7077 | 3282 | 2080 | 8.7 |
| Ficedula albicollis | 15 922 | 16 148 | 64 | 985 | 390 | 57 | 778 | 4669 | 81 | 5208 | 2021 | 1049 | 6.5 |
| Taeniopygia guttata | 18 141 | 19 724 | 85 | 716 | 432 | 77 | 972 | 6749 | 72 | 6197 | 2211 | 801 | 4.1 |
| Chelonia mydas | 19 066 | 71 | 880 | 478 | 71 | 794 | 6031 | 97 | 6384 | 2194 | 951 | 5.0 | |
| Pelodiscus sinensis | 20 784 | 126 | 1271 | 492 | 64 | 798 | 6724 | 92 | 6703 | 2683 | 1397 | 6.7 | |
| Other vertebrata | |||||||||||||
| Petromyzon marinus | 13 160 | 54 | 522 | 255 | 66 | 669 | 3945 | 34 | 3797 | 1239 | 576 | 4.4 | |
| Latimeria chalumnae | 23 429 | 23 513 | 75 | 1270 | 593 | 87 | 993 | 8117 | 116 | 7349 | 2905 | 1345 | 5.7 |
| Anolis carolinensis | 19 109 | 19 562 | 81 | 1238 | 510 | 288 | 936 | 5727 | 89 | 6435 | 2478 | 1319 | 6.7 |
| Invertebrate | |||||||||||||
| Chordata (Tunicata) | |||||||||||||
| Oikopleura dioica* | 17 050 | 29 057 | 15 | 1864 | 1177 | 116 | 1493 | 10 785 | 92 | 8060 | 2540 | 1879 | 6.5 |
| Ciona intestinalis | 17 308 | 18 639 | 28 | 1507 | 601 | 95 | 738 | 6231 | 47 | 5014 | 1812 | 1535 | 8.2 |
| C. savignyi | 20 004 | 20 117 | 45 | 822 | 359 | 52 | 697 | 7792 | 55 | 5685 | 2535 | 867 | 4.3 |
| Ecdysozoa (Arachnida) | |||||||||||||
| Tetranychus urticae | 18 082 | 18 243 | 12 | 1891 | 685 | 95 | 701 | 5986 | 59 | 3615 | 2219 | 1903 | 10.4 |
| Ixodes ricinus | 16 199 | 1 | 3554 | 819 | 136 | 848 | 3528 | 97 | 3649 | 1246 | 3555 | 21.9 | |
| I. scapularis | 20 473 | 21 162 | 21 | 1943 | 762 | 103 | 1433 | 5706 | 95 | 5905 | 1681 | 1964 | 9.3 |
| Rhipicephalus pulchellus | 11 205 | 1 | 1620 | 459 | 60 | 962 | 2380 | 49 | 3424 | 1231 | 1621 | 14.5 | |
| Ecdysozoa (Insecta) | |||||||||||||
| Drosophila mojavensis | 14 525 | 15 086 | 21 | 2049 | 347 | 95 | 832 | 4302 | 74 | 4241 | 1700 | 2070 | 13.7 |
| D. virilis | 14 456 | 14 928 | 34 | 1941 | 354 | 84 | 770 | 4308 | 57 | 4323 | 1709 | 1975 | 13.2 |
| D. erecta | 15 116 | 44 | 2220 | 362 | 71 | 873 | 3918 | 59 | 4366 | 1721 | 2264 | 15.0 | |
| D. grimshawi | 14 754 | 14 798 | 31 | 1861 | 362 | 62 | 773 | 4336 | 79 | 4195 | 1698 | 1892 | 12.8 |
| D. ananassae | 14 968 | 15 298 | 28 | 2139 | 349 | 64 | 791 | 4243 | 67 | 4503 | 1793 | 2167 | 14.2 |
| D. melanogaster* | 20 120 | 39 951 | 254 | 4761 | 923 | 269 | 2127 | 10 613 | 191 | 12 101 | 4659 | 5015 | 12.6 |
| D. persimilis | 16 754 | 16 861 | 28 | 2106 | 420 | 77 | 930 | 4846 | 68 | 5076 | 1701 | 2134 | 12.7 |
| D. pseudoobscura pseudoobscura | 17 047 | 48 | 2316 | 416 | 77 | 939 | 4632 | 71 | 5126 | 1950 | 2364 | 13.9 | |
| D. sechellia | 16 134 | 16 361 | 37 | 2250 | 410 | 71 | 936 | 4464 | 54 | 4765 | 1734 | 2287 | 14.0 |
| D. simulans | 15 354 | 19 057 | 57 | 2372 | 436 | 100 | 1028 | 5483 | 56 | 5374 | 2165 | 2429 | 12.7 |
| D. willistoni | 15 447 | 15 564 | 25 | 1875 | 355 | 77 | 815 | 4808 | 60 | 4434 | 1722 | 1900 | 12.2 |
| D. yakuba | 17 257 | 41 | 2521 | 392 | 77 | 1006 | 4752 | 59 | 5024 | 1798 | 2562 | 14.8 | |
| Megaselia scalaris | 11 463 | 11 503 | 10 | 773 | 417 | 31 | 449 | 4947 | 24 | 2402 | 639 | 783 | 6.8 |
| Anopheles darlingi | 10 447 | 11 686 | 8 | 793 | 272 | 93 | 758 | 3687 | 54 | 3971 | 1162 | 801 | 6.9 |
| A. gambiae* | 13 072 | 19 384 | 50 | 2610 | 410 | 87 | 1104 | 6027 | 57 | 4834 | 1869 | 2660 | 13.7 |
| Aedes aegypti | 16 654 | 17 683 | 54 | 2367 | 469 | 182 | 961 | 5052 | 56 | 4873 | 2008 | 2421 | 13.7 |
| Culex quinquefasciatus | 18 703 | 19 062 | 25 | 2345 | 534 | 128 | 1104 | 5751 | 73 | 5501 | 1866 | 2370 | 12.4 |
| Dendroctonus ponderosae | 23 650 | 14 | 1992 | 549 | 106 | 1153 | 8928 | 96 | 6087 | 2502 | 2006 | 8.5 | |
| Tribolium castaneum | 16 502 | 17 074 | 26 | 1717 | 423 | 66 | 846 | 5830 | 50 | 4196 | 2109 | 1743 | 10.2 |
| Apis mellifera | 10 910 | 12 299 | 65 | 757 | 267 | 81 | 390 | 4440 | 45 | 3293 | 1714 | 822 | 6.7 |
| Camponotus floridanus | 14 787 | 14 801 | 15 | 662 | 329 | 45 | 744 | 5533 | 65 | 4078 | 1346 | 677 | 4.6 |
| Acromyrmex echinatior | 13 962 | 13 970 | 17 | 592 | 327 | 36 | 847 | 5226 | 62 | 4219 | 1253 | 609 | 4.4 |
| Atta cephalotes | 18 079 | 18 113 | 16 | 753 | 597 | 99 | 1094 | 6579 | 75 | 4715 | 1559 | 769 | 4.2 |
| Solenopsis invicta | 14 193 | 14 359 | 26 | 636 | 437 | 100 | 748 | 5413 | 31 | 3508 | 1120 | 662 | 4.6 |
| Harpegnathos saltator | 15 029 | 15 042 | 17 | 739 | 329 | 46 | 696 | 5484 | 60 | 4223 | 1299 | 756 | 5.0 |
| Nasonia vitripennis | 17 040 | 17 289 | 14 | 1545 | 305 | 65 | 701 | 6951 | 55 | 4883 | 1423 | 1559 | 9.0 |
| Bombyx mori | 14 767 | 17 915 | 125 | 1773 | 379 | 108 | 806 | 6293 | 54 | 4580 | 1681 | 1898 | 10.6 |
| Danaus plexippus | 16 253 | 16 358 | 34 | 1486 | 441 | 95 | 808 | 5657 | 66 | 4528 | 1493 | 1520 | 9.3 |
| Pararge aegeria | 15 104 | 12 | 428 | 561 | 75 | 850 | 5983 | 14 | 3763 | 503 | 440 | 2.9 | |
| Rhodnius prolixus | 15 180 | 16 639 | 44 | 1420 | 537 | 41 | 562 | 6782 | 62 | 3769 | 1473 | 1464 | 8.8 |
| Acyrthosiphon pisum | 35 809 | 35 211 | 24 | 1834 | 814 | 102 | 1736 | 15 209 | 66 | 8622 | 1736 | 1858 | 5.3 |
| Pediculus humanus subsp. corporis | 10 847 | 11 | 513 | 257 | 37 | 349 | 4294 | 40 | 3174 | 1193 | 524 | 4.8 | |
| Ecdysozoa (Nematoda) | |||||||||||||
| Ascaris suum* | 9213 | 18 539 | 39 | 1223 | 577 | 107 | 1302 | 5894 | 75 | 4965 | 2437 | 1262 | 6.8 |
| Pristionchus pacificus | 29 079 | 29 319 | 14 | 3027 | 1038 | 75 | 1368 | 9699 | 94 | 7157 | 3263 | 3041 | 10.4 |
| Caenorhabditis brenneri | 29 982 | 30 712 | 21 | 3602 | 896 | 76 | 1134 | 10 314 | 88 | 7338 | 4255 | 3623 | 11.8 |
| C. briggsae | 21 751 | 21 914 | 30 | 2734 | 655 | 119 | 874 | 6435 | 106 | 5178 | 3540 | 2764 | 12.6 |
| C. elegans | 26 173 | 26 447 | 182 | 3573 | 856 | 163 | 1065 | 7156 | 107 | 6190 | 4401 | 3755 | 14.2 |
| C. japonica | 35 063 | 35 069 | 14 | 2665 | 998 | 95 | 2061 | 12 267 | 123 | 9112 | 3234 | 2679 | 7.6 |
| C. remanei | 31 252 | 32 133 | 21 | 3859 | 1117 | 84 | 1282 | 10 352 | 93 | 7199 | 4981 | 3880 | 12.1 |
| Haemonchus contortus | 18 580 | 3 | 2181 | 558 | 83 | 1016 | 5826 | 77 | 4679 | 2261 | 2184 | 11.8 | |
| Brugia malayi* | 1643 | 11 561 | 10 | 668 | 347 | 37 | 579 | 4540 | 44 | 3139 | 908 | 678 | 5.9 |
| Loa loa | 15 319 | 15 356 | 11 | 784 | 588 | 46 | 750 | 5774 | 49 | 3749 | 1387 | 795 | 5.2 |
| Wuchereria bancrofti | 19 298 | 19 525 | 18 | 716 | 677 | 129 | 870 | 8254 | 39 | 4504 | 1205 | 734 | 3.8 |
| Trichinella spiralis | 16 041 | 16 278 | 17 | 935 | 770 | 73 | 980 | 5389 | 71 | 3433 | 1234 | 952 | 5.8 |
| Ecdysozoa (Arthropoda) | |||||||||||||
| Daphnia pulex | 30 137 | 30 988 | 22 | 2827 | 892 | 333 | 1432 | 11 433 | 88 | 8315 | 2130 | 2849 | 9.2 |
| Strigamia maritima | 14 972 | 15 011 | 19 | 1118 | 428 | 48 | 726 | 5331 | 68 | 3635 | 1835 | 1137 | 7.6 |
| Lophotrochozoa | |||||||||||||
| Helobdella robusta | 23 328 | 23 379 | 19 | 1170 | 671 | 59 | 924 | 9385 | 145 | 5866 | 2226 | 1189 | 5.1 |
| Capitella teleta | 31 207 | 22 | 2183 | 907 | 76 | 1263 | 10 827 | 106 | 7765 | 3917 | 2205 | 7.1 | |
| Crassostrea gigas | 25 982 | 26 850 | 26 | 1904 | 633 | 85 | 814 | 10 178 | 140 | 7045 | 2912 | 1930 | 7.2 |
| Lottia gigantea | 23 721 | 34 | 1683 | 588 | 48 | 659 | 9382 | 76 | 5530 | 2734 | 1717 | 7.2 | |
| Platyhelminthes | |||||||||||||
| Echinococcus granulosus | 11 124 | 0 | 614 | 375 | 381 | 656 | 2855 | 40 | 3518 | 1260 | 614 | 5.5 | |
| E. multilocularis | 10 572 | 0 | 591 | 326 | 91 | 656 | 2878 | 48 | 3532 | 1239 | 591 | 5.6 | |
| Clonorchis sinensis | 13 606 | 13 880 | 6 | 562 | 349 | 55 | 990 | 4294 | 89 | 5074 | 1234 | 568 | 4.1 |
| Schistosoma japonicum | 16 236 | 17 | 1767 | 607 | 70 | 853 | 6086 | 36 | 3511 | 1357 | 1784 | 11.0 | |
| S. mansoni | 11 723 | 12 836 | 9 | 605 | 427 | 203 | 505 | 4491 | 60 | 3740 | 1242 | 614 | 4.8 |
| Other Invertebrates | |||||||||||||
| Amphimedon queenslandica | 29 741 | 29 816 | 6 | 1490 | 893 | 65 | 1246 | 11 722 | 73 | 7333 | 2685 | 1496 | 5.0 |
| Nematostella vectensis | 24 435 | 25 035 | 61 | 1135 | 586 | 58 | 1005 | 8651 | 72 | 6385 | 3293 | 1196 | 4.8 |
| Strongylocentrotus purpuratus | 28 567 | 29 560 | 46 | 2198 | 737 | 94 | 1101 | 9895 | 145 | 8580 | 4026 | 2244 | 7.6 |
| Trichoplax adhaerens | 11 520 | 11 590 | 7 | 482 | 213 | 36 | 489 | 5013 | 44 | 2502 | 1776 | 489 | 4.2 |
| Branchiostoma floridae | 28 544 | 29 237 | 37 | 2227 | 710 | 152 | 1146 | 8799 | 140 | 7800 | 3826 | 2264 | 7.7 |
Notes : Data of other protein subcellular locations are summarized in Supplementary Table 1 . HLS: highly likely secreted; LS: likely secreted; Mito: mitochondrial; mem: membrane; non-mem: non-membrane; Cyt: cytoplasm (or cytosol); Nuc: nuclear; Secr: secretome. Species labeled with * has more (or less) 5000 protein entries than its reference proteome.
Summary of proteins located in some major subcellular locations in human and different animal species
| . | Referenceproteome . | Totalproteins . | Curatedsecreted . | Predicted . | Mito . | . | Nuc . | Plasmamem . | . | Secr(%) . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | HLS . | LS . | mem . | non-mem . | Cyt . | mem . | non-mem . | . | Secr . | . | |
| Vertebrata (Actinopterygii) | |||||||||||||
| Oryzias latipes | 24 633 | 26 060 | 144 | 1805 | 649 | 141 | 1185 | 7330 | 162 | 8629 | 3580 | 1949 | 7.5 |
| Xiphophorus maculatus | 20 451 | 20 527 | 92 | 1476 | 510 | 73 | 959 | 5288 | 92 | 7237 | 3052 | 1568 | 7.6 |
| Oreochromis niloticus | 26 753 | 27 551 | 122 | 2179 | 638 | 148 | 1051 | 6971 | 149 | 9638 | 4078 | 2301 | 8.4 |
| Gasterosteus aculeatus | 27 248 | 28 110 | 114 | 1813 | 618 | 106 | 1418 | 8080 | 142 | 9443 | 3877 | 1927 | 6.9 |
| Takifugu rubripes | 47 856 | 49 090 | 261 | 2655 | 1028 | 172 | 1645 | 12 630 | 323 | 17 843 | 8443 | 2916 | 5.9 |
| Tetraodon nigroviridis* | 23 073 | 49 327 | 194 | 2700 | 1236 | 182 | 2248 | 13 333 | 337 | 16 961 | 5944 | 2894 | 5.9 |
| Danio rerio* | 41 054 | 55 414 | 372 | 4635 | 1189 | 282 | 2319 | 14 909 | 267 | 19 521 | 6866 | 5007 | 9.0 |
| Vertebrata (Amphibia) | |||||||||||||
| Xenopus tropicalis* | 23 491 | 30 521 | 194 | 1926 | 656 | 169 | 1327 | 9674 | 168 | 10 086 | 4113 | 2120 | 6.9 |
| X. laevis | 16 011 | 269 | 1059 | 262 | 161 | 752 | 5124 | 109 | 5711 | 1636 | 1328 | 8.3 | |
| Vertebrata (Mammalia) | |||||||||||||
| Glires | |||||||||||||
| Oryctolagus cuniculus | 21 150 | 22 788 | 334 | 1670 | 479 | 135 | 1222 | 5692 | 150 | 7241 | 3665 | 2004 | 8.8 |
| Heterocephalus glaber | 21 449 | 21 548 | 93 | 1266 | 513 | 90 | 1009 | 6343 | 103 | 6924 | 3266 | 1359 | 6.3 |
| Cavia porcellus | 19 911 | 20 378 | 236 | 1432 | 461 | 100 | 1016 | 5349 | 103 | 6410 | 3342 | 1668 | 8.2 |
| Cricetulus griseus | 23 884 | 24 442 | 109 | 1407 | 927 | 96 | 1170 | 7073 | 116 | 7114 | 2786 | 1516 | 6.2 |
| Mus musculus* | 43 539 | 74 158 | 1792 | 4350 | 1698 | 717 | 3443 | 20 456 | 714 | 23 226 | 9137 | 6142 | 8.3 |
| Rattus norvegicus* | 27 340 | 33 555 | 966 | 2211 | 637 | 476 | 1473 | 9153 | 411 | 10 094 | 5407 | 3177 | 9.5 |
| Spermophilus tridecemlineatus | 19 966 | 20 079 | 110 | 1437 | 429 | 83 | 937 | 5488 | 103 | 6603 | 3290 | 1547 | 7.7 |
| Primates | |||||||||||||
| Macaca fascicularis * | 17 396 | 28 955 | 233 | 1912 | 928 | 359 | 1976 | 7639 | 121 | 8186 | 2970 | 2145 | 7.4 |
| M. mulatta* | 35 536 | 69 567 | 407 | 4554 | 1694 | 653 | 3719 | 18 667 | 326 | 23 502 | 7295 | 4961 | 7.1 |
| Gorilla gorilla gorilla | 27 286 | 27 371 | 212 | 1994 | 676 | 218 | 1480 | 6701 | 151 | 9481 | 3358 | 2206 | 8.1 |
| Homo sapiens* | 68 049 | 13 5661 | 2020 | 6682 | 3480 | 3737 | 17 623 | 34 825 | 877 | 34 274 | 10 607 | 8702 | 6.4 |
| Pan troglodytes* | 20 126 | 33 326 | 296 | 2241 | 820 | 447 | 1825 | 7966 | 137 | 11 618 | 4190 | 2537 | 7.6 |
| Pongo abelii | 22 785 | 24 529 | 237 | 1818 | 580 | 229 | 1457 | 6452 | 168 | 8228 | 2879 | 2055 | 8.4 |
| Nomascus leucogenys | 19 734 | 19 837 | 141 | 1489 | 518 | 114 | 1143 | 5053 | 99 | 6893 | 2457 | 1630 | 8.2 |
| Callithrix jacchus* | 42 025 | 55 085 | 244 | 3776 | 1280 | 195 | 2867 | 15 064 | 308 | 20 159 | 6178 | 4020 | 7.3 |
| Otolemur garnettii | 19 930 | 20 156 | 99 | 1515 | 480 | 93 | 1022 | 5226 | 96 | 6801 | 3099 | 1614 | 8.0 |
| Carnivora | |||||||||||||
| Canis familiaris | 25 439 | 28 362 | 345 | 1813 | 595 | 385 | 1491 | 7040 | 170 | 9489 | 4047 | 2158 | 7.6 |
| Mustela putorius furo | 38 826 | 173 | 2017 | 984 | 137 | 2127 | 11 785 | 154 | 12 830 | 4073 | 2190 | 5.6 | |
| Neovison vison | 16 237 | 18 | 750 | 356 | 66 | 839 | 5636 | 52 | 5233 | 1507 | 768 | 4.7 | |
| Ailuropoda melanoleuca* | 21 136 | 35 743 | 247 | 2086 | 779 | 176 | 1746 | 9975 | 162 | 11 905 | 5133 | 2333 | 6.5 |
| Felis catus | 20 303 | 21 230 | 196 | 1406 | 483 | 108 | 1065 | 5831 | 107 | 6791 | 3091 | 1602 | 7.5 |
| Cetartiodactyla | |||||||||||||
| Bos mutus | 18 922 | 150 | 1377 | 453 | 123 | 931 | 4911 | 85 | 5854 | 3159 | 1527 | 8.1 | |
| Bos taurus* | 23 842 | 31 780 | 880 | 2215 | 620 | 508 | 1491 | 8171 | 317 | 9598 | 4492 | 3095 | 9.7 |
| Sus scrofa* | 26 054 | 33 962 | 645 | 2534 | 779 | 411 | 1560 | 9038 | 166 | 9463 | 4787 | 3179 | 9.4 |
| Camelus ferus | 20 028 | 67 | 1084 | 636 | 99 | 1132 | 5715 | 147 | 6257 | 2588 | 1151 | 5.7 | |
| Chiroptera | |||||||||||||
| Pteropus alecto | 19 520 | 19 548 | 97 | 1162 | 488 | 74 | 1160 | 5364 | 121 | 6774 | 2447 | 1259 | 6.4 |
| Myotis brandtii | 19 301 | 58 | 1032 | 432 | 90 | 938 | 5806 | 104 | 6427 | 2250 | 1090 | 5.6 | |
| M. davidii | 15 446 | 15 466 | 67 | 916 | 345 | 60 | 782 | 4530 | 73 | 5194 | 1816 | 983 | 6.4 |
| M. lucifugus | 20 650 | 20 899 | 143 | 1738 | 431 | 100 | 1052 | 5716 | 96 | 6782 | 2855 | 1881 | 9.0 |
| Other mammalia | |||||||||||||
| Loxodonta africana | 25 615 | 25 832 | 132 | 1744 | 556 | 128 | 1119 | 6554 | 129 | 8459 | 4835 | 1876 | 7.3 |
| Equus caballus | 22 676 | 27 841 | 272 | 1659 | 514 | 284 | 1042 | 8886 | 133 | 8701 | 3825 | 1931 | 6.9 |
| Tupaia chinensis | 20 824 | 20 851 | 85 | 1275 | 527 | 64 | 1149 | 5699 | 125 | 6701 | 3114 | 1360 | 6.5 |
| Sarcophilus harrisii | 22 388 | 22 565 | 107 | 1490 | 553 | 110 | 867 | 6368 | 102 | 7495 | 3495 | 1597 | 7.1 |
| Monodelphis domestica | 22 240 | 22 794 | 108 | 1505 | 485 | 84 | 1103 | 6398 | 106 | 7252 | 3930 | 1613 | 7.1 |
| Ornithorhynchus anatinus | 23 552 | 23 763 | 113 | 1202 | 698 | 103 | 1111 | 7229 | 95 | 7184 | 3157 | 1315 | 5.5 |
| Vertebrata (Testudines + Archosauria group) | |||||||||||||
| Anas platyrhynchos * | 16 377 | 31 879 | 139 | 1360 | 893 | 123 | 1269 | 10 542 | 148 | 9829 | 3316 | 1499 | 4.7 |
| Meleagris gallopavo | 16 537 | 16 991 | 114 | 892 | 413 | 75 | 673 | 5622 | 83 | 5377 | 2073 | 1006 | 5.9 |
| Gallus gallus* | 17 623 | 23 800 | 440 | 1640 | 581 | 278 | 1231 | 6403 | 147 | 7077 | 3282 | 2080 | 8.7 |
| Ficedula albicollis | 15 922 | 16 148 | 64 | 985 | 390 | 57 | 778 | 4669 | 81 | 5208 | 2021 | 1049 | 6.5 |
| Taeniopygia guttata | 18 141 | 19 724 | 85 | 716 | 432 | 77 | 972 | 6749 | 72 | 6197 | 2211 | 801 | 4.1 |
| Chelonia mydas | 19 066 | 71 | 880 | 478 | 71 | 794 | 6031 | 97 | 6384 | 2194 | 951 | 5.0 | |
| Pelodiscus sinensis | 20 784 | 126 | 1271 | 492 | 64 | 798 | 6724 | 92 | 6703 | 2683 | 1397 | 6.7 | |
| Other vertebrata | |||||||||||||
| Petromyzon marinus | 13 160 | 54 | 522 | 255 | 66 | 669 | 3945 | 34 | 3797 | 1239 | 576 | 4.4 | |
| Latimeria chalumnae | 23 429 | 23 513 | 75 | 1270 | 593 | 87 | 993 | 8117 | 116 | 7349 | 2905 | 1345 | 5.7 |
| Anolis carolinensis | 19 109 | 19 562 | 81 | 1238 | 510 | 288 | 936 | 5727 | 89 | 6435 | 2478 | 1319 | 6.7 |
| Invertebrate | |||||||||||||
| Chordata (Tunicata) | |||||||||||||
| Oikopleura dioica* | 17 050 | 29 057 | 15 | 1864 | 1177 | 116 | 1493 | 10 785 | 92 | 8060 | 2540 | 1879 | 6.5 |
| Ciona intestinalis | 17 308 | 18 639 | 28 | 1507 | 601 | 95 | 738 | 6231 | 47 | 5014 | 1812 | 1535 | 8.2 |
| C. savignyi | 20 004 | 20 117 | 45 | 822 | 359 | 52 | 697 | 7792 | 55 | 5685 | 2535 | 867 | 4.3 |
| Ecdysozoa (Arachnida) | |||||||||||||
| Tetranychus urticae | 18 082 | 18 243 | 12 | 1891 | 685 | 95 | 701 | 5986 | 59 | 3615 | 2219 | 1903 | 10.4 |
| Ixodes ricinus | 16 199 | 1 | 3554 | 819 | 136 | 848 | 3528 | 97 | 3649 | 1246 | 3555 | 21.9 | |
| I. scapularis | 20 473 | 21 162 | 21 | 1943 | 762 | 103 | 1433 | 5706 | 95 | 5905 | 1681 | 1964 | 9.3 |
| Rhipicephalus pulchellus | 11 205 | 1 | 1620 | 459 | 60 | 962 | 2380 | 49 | 3424 | 1231 | 1621 | 14.5 | |
| Ecdysozoa (Insecta) | |||||||||||||
| Drosophila mojavensis | 14 525 | 15 086 | 21 | 2049 | 347 | 95 | 832 | 4302 | 74 | 4241 | 1700 | 2070 | 13.7 |
| D. virilis | 14 456 | 14 928 | 34 | 1941 | 354 | 84 | 770 | 4308 | 57 | 4323 | 1709 | 1975 | 13.2 |
| D. erecta | 15 116 | 44 | 2220 | 362 | 71 | 873 | 3918 | 59 | 4366 | 1721 | 2264 | 15.0 | |
| D. grimshawi | 14 754 | 14 798 | 31 | 1861 | 362 | 62 | 773 | 4336 | 79 | 4195 | 1698 | 1892 | 12.8 |
| D. ananassae | 14 968 | 15 298 | 28 | 2139 | 349 | 64 | 791 | 4243 | 67 | 4503 | 1793 | 2167 | 14.2 |
| D. melanogaster* | 20 120 | 39 951 | 254 | 4761 | 923 | 269 | 2127 | 10 613 | 191 | 12 101 | 4659 | 5015 | 12.6 |
| D. persimilis | 16 754 | 16 861 | 28 | 2106 | 420 | 77 | 930 | 4846 | 68 | 5076 | 1701 | 2134 | 12.7 |
| D. pseudoobscura pseudoobscura | 17 047 | 48 | 2316 | 416 | 77 | 939 | 4632 | 71 | 5126 | 1950 | 2364 | 13.9 | |
| D. sechellia | 16 134 | 16 361 | 37 | 2250 | 410 | 71 | 936 | 4464 | 54 | 4765 | 1734 | 2287 | 14.0 |
| D. simulans | 15 354 | 19 057 | 57 | 2372 | 436 | 100 | 1028 | 5483 | 56 | 5374 | 2165 | 2429 | 12.7 |
| D. willistoni | 15 447 | 15 564 | 25 | 1875 | 355 | 77 | 815 | 4808 | 60 | 4434 | 1722 | 1900 | 12.2 |
| D. yakuba | 17 257 | 41 | 2521 | 392 | 77 | 1006 | 4752 | 59 | 5024 | 1798 | 2562 | 14.8 | |
| Megaselia scalaris | 11 463 | 11 503 | 10 | 773 | 417 | 31 | 449 | 4947 | 24 | 2402 | 639 | 783 | 6.8 |
| Anopheles darlingi | 10 447 | 11 686 | 8 | 793 | 272 | 93 | 758 | 3687 | 54 | 3971 | 1162 | 801 | 6.9 |
| A. gambiae* | 13 072 | 19 384 | 50 | 2610 | 410 | 87 | 1104 | 6027 | 57 | 4834 | 1869 | 2660 | 13.7 |
| Aedes aegypti | 16 654 | 17 683 | 54 | 2367 | 469 | 182 | 961 | 5052 | 56 | 4873 | 2008 | 2421 | 13.7 |
| Culex quinquefasciatus | 18 703 | 19 062 | 25 | 2345 | 534 | 128 | 1104 | 5751 | 73 | 5501 | 1866 | 2370 | 12.4 |
| Dendroctonus ponderosae | 23 650 | 14 | 1992 | 549 | 106 | 1153 | 8928 | 96 | 6087 | 2502 | 2006 | 8.5 | |
| Tribolium castaneum | 16 502 | 17 074 | 26 | 1717 | 423 | 66 | 846 | 5830 | 50 | 4196 | 2109 | 1743 | 10.2 |
| Apis mellifera | 10 910 | 12 299 | 65 | 757 | 267 | 81 | 390 | 4440 | 45 | 3293 | 1714 | 822 | 6.7 |
| Camponotus floridanus | 14 787 | 14 801 | 15 | 662 | 329 | 45 | 744 | 5533 | 65 | 4078 | 1346 | 677 | 4.6 |
| Acromyrmex echinatior | 13 962 | 13 970 | 17 | 592 | 327 | 36 | 847 | 5226 | 62 | 4219 | 1253 | 609 | 4.4 |
| Atta cephalotes | 18 079 | 18 113 | 16 | 753 | 597 | 99 | 1094 | 6579 | 75 | 4715 | 1559 | 769 | 4.2 |
| Solenopsis invicta | 14 193 | 14 359 | 26 | 636 | 437 | 100 | 748 | 5413 | 31 | 3508 | 1120 | 662 | 4.6 |
| Harpegnathos saltator | 15 029 | 15 042 | 17 | 739 | 329 | 46 | 696 | 5484 | 60 | 4223 | 1299 | 756 | 5.0 |
| Nasonia vitripennis | 17 040 | 17 289 | 14 | 1545 | 305 | 65 | 701 | 6951 | 55 | 4883 | 1423 | 1559 | 9.0 |
| Bombyx mori | 14 767 | 17 915 | 125 | 1773 | 379 | 108 | 806 | 6293 | 54 | 4580 | 1681 | 1898 | 10.6 |
| Danaus plexippus | 16 253 | 16 358 | 34 | 1486 | 441 | 95 | 808 | 5657 | 66 | 4528 | 1493 | 1520 | 9.3 |
| Pararge aegeria | 15 104 | 12 | 428 | 561 | 75 | 850 | 5983 | 14 | 3763 | 503 | 440 | 2.9 | |
| Rhodnius prolixus | 15 180 | 16 639 | 44 | 1420 | 537 | 41 | 562 | 6782 | 62 | 3769 | 1473 | 1464 | 8.8 |
| Acyrthosiphon pisum | 35 809 | 35 211 | 24 | 1834 | 814 | 102 | 1736 | 15 209 | 66 | 8622 | 1736 | 1858 | 5.3 |
| Pediculus humanus subsp. corporis | 10 847 | 11 | 513 | 257 | 37 | 349 | 4294 | 40 | 3174 | 1193 | 524 | 4.8 | |
| Ecdysozoa (Nematoda) | |||||||||||||
| Ascaris suum* | 9213 | 18 539 | 39 | 1223 | 577 | 107 | 1302 | 5894 | 75 | 4965 | 2437 | 1262 | 6.8 |
| Pristionchus pacificus | 29 079 | 29 319 | 14 | 3027 | 1038 | 75 | 1368 | 9699 | 94 | 7157 | 3263 | 3041 | 10.4 |
| Caenorhabditis brenneri | 29 982 | 30 712 | 21 | 3602 | 896 | 76 | 1134 | 10 314 | 88 | 7338 | 4255 | 3623 | 11.8 |
| C. briggsae | 21 751 | 21 914 | 30 | 2734 | 655 | 119 | 874 | 6435 | 106 | 5178 | 3540 | 2764 | 12.6 |
| C. elegans | 26 173 | 26 447 | 182 | 3573 | 856 | 163 | 1065 | 7156 | 107 | 6190 | 4401 | 3755 | 14.2 |
| C. japonica | 35 063 | 35 069 | 14 | 2665 | 998 | 95 | 2061 | 12 267 | 123 | 9112 | 3234 | 2679 | 7.6 |
| C. remanei | 31 252 | 32 133 | 21 | 3859 | 1117 | 84 | 1282 | 10 352 | 93 | 7199 | 4981 | 3880 | 12.1 |
| Haemonchus contortus | 18 580 | 3 | 2181 | 558 | 83 | 1016 | 5826 | 77 | 4679 | 2261 | 2184 | 11.8 | |
| Brugia malayi* | 1643 | 11 561 | 10 | 668 | 347 | 37 | 579 | 4540 | 44 | 3139 | 908 | 678 | 5.9 |
| Loa loa | 15 319 | 15 356 | 11 | 784 | 588 | 46 | 750 | 5774 | 49 | 3749 | 1387 | 795 | 5.2 |
| Wuchereria bancrofti | 19 298 | 19 525 | 18 | 716 | 677 | 129 | 870 | 8254 | 39 | 4504 | 1205 | 734 | 3.8 |
| Trichinella spiralis | 16 041 | 16 278 | 17 | 935 | 770 | 73 | 980 | 5389 | 71 | 3433 | 1234 | 952 | 5.8 |
| Ecdysozoa (Arthropoda) | |||||||||||||
| Daphnia pulex | 30 137 | 30 988 | 22 | 2827 | 892 | 333 | 1432 | 11 433 | 88 | 8315 | 2130 | 2849 | 9.2 |
| Strigamia maritima | 14 972 | 15 011 | 19 | 1118 | 428 | 48 | 726 | 5331 | 68 | 3635 | 1835 | 1137 | 7.6 |
| Lophotrochozoa | |||||||||||||
| Helobdella robusta | 23 328 | 23 379 | 19 | 1170 | 671 | 59 | 924 | 9385 | 145 | 5866 | 2226 | 1189 | 5.1 |
| Capitella teleta | 31 207 | 22 | 2183 | 907 | 76 | 1263 | 10 827 | 106 | 7765 | 3917 | 2205 | 7.1 | |
| Crassostrea gigas | 25 982 | 26 850 | 26 | 1904 | 633 | 85 | 814 | 10 178 | 140 | 7045 | 2912 | 1930 | 7.2 |
| Lottia gigantea | 23 721 | 34 | 1683 | 588 | 48 | 659 | 9382 | 76 | 5530 | 2734 | 1717 | 7.2 | |
| Platyhelminthes | |||||||||||||
| Echinococcus granulosus | 11 124 | 0 | 614 | 375 | 381 | 656 | 2855 | 40 | 3518 | 1260 | 614 | 5.5 | |
| E. multilocularis | 10 572 | 0 | 591 | 326 | 91 | 656 | 2878 | 48 | 3532 | 1239 | 591 | 5.6 | |
| Clonorchis sinensis | 13 606 | 13 880 | 6 | 562 | 349 | 55 | 990 | 4294 | 89 | 5074 | 1234 | 568 | 4.1 |
| Schistosoma japonicum | 16 236 | 17 | 1767 | 607 | 70 | 853 | 6086 | 36 | 3511 | 1357 | 1784 | 11.0 | |
| S. mansoni | 11 723 | 12 836 | 9 | 605 | 427 | 203 | 505 | 4491 | 60 | 3740 | 1242 | 614 | 4.8 |
| Other Invertebrates | |||||||||||||
| Amphimedon queenslandica | 29 741 | 29 816 | 6 | 1490 | 893 | 65 | 1246 | 11 722 | 73 | 7333 | 2685 | 1496 | 5.0 |
| Nematostella vectensis | 24 435 | 25 035 | 61 | 1135 | 586 | 58 | 1005 | 8651 | 72 | 6385 | 3293 | 1196 | 4.8 |
| Strongylocentrotus purpuratus | 28 567 | 29 560 | 46 | 2198 | 737 | 94 | 1101 | 9895 | 145 | 8580 | 4026 | 2244 | 7.6 |
| Trichoplax adhaerens | 11 520 | 11 590 | 7 | 482 | 213 | 36 | 489 | 5013 | 44 | 2502 | 1776 | 489 | 4.2 |
| Branchiostoma floridae | 28 544 | 29 237 | 37 | 2227 | 710 | 152 | 1146 | 8799 | 140 | 7800 | 3826 | 2264 | 7.7 |
| . | Referenceproteome . | Totalproteins . | Curatedsecreted . | Predicted . | Mito . | . | Nuc . | Plasmamem . | . | Secr(%) . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | HLS . | LS . | mem . | non-mem . | Cyt . | mem . | non-mem . | . | Secr . | . | |
| Vertebrata (Actinopterygii) | |||||||||||||
| Oryzias latipes | 24 633 | 26 060 | 144 | 1805 | 649 | 141 | 1185 | 7330 | 162 | 8629 | 3580 | 1949 | 7.5 |
| Xiphophorus maculatus | 20 451 | 20 527 | 92 | 1476 | 510 | 73 | 959 | 5288 | 92 | 7237 | 3052 | 1568 | 7.6 |
| Oreochromis niloticus | 26 753 | 27 551 | 122 | 2179 | 638 | 148 | 1051 | 6971 | 149 | 9638 | 4078 | 2301 | 8.4 |
| Gasterosteus aculeatus | 27 248 | 28 110 | 114 | 1813 | 618 | 106 | 1418 | 8080 | 142 | 9443 | 3877 | 1927 | 6.9 |
| Takifugu rubripes | 47 856 | 49 090 | 261 | 2655 | 1028 | 172 | 1645 | 12 630 | 323 | 17 843 | 8443 | 2916 | 5.9 |
| Tetraodon nigroviridis* | 23 073 | 49 327 | 194 | 2700 | 1236 | 182 | 2248 | 13 333 | 337 | 16 961 | 5944 | 2894 | 5.9 |
| Danio rerio* | 41 054 | 55 414 | 372 | 4635 | 1189 | 282 | 2319 | 14 909 | 267 | 19 521 | 6866 | 5007 | 9.0 |
| Vertebrata (Amphibia) | |||||||||||||
| Xenopus tropicalis* | 23 491 | 30 521 | 194 | 1926 | 656 | 169 | 1327 | 9674 | 168 | 10 086 | 4113 | 2120 | 6.9 |
| X. laevis | 16 011 | 269 | 1059 | 262 | 161 | 752 | 5124 | 109 | 5711 | 1636 | 1328 | 8.3 | |
| Vertebrata (Mammalia) | |||||||||||||
| Glires | |||||||||||||
| Oryctolagus cuniculus | 21 150 | 22 788 | 334 | 1670 | 479 | 135 | 1222 | 5692 | 150 | 7241 | 3665 | 2004 | 8.8 |
| Heterocephalus glaber | 21 449 | 21 548 | 93 | 1266 | 513 | 90 | 1009 | 6343 | 103 | 6924 | 3266 | 1359 | 6.3 |
| Cavia porcellus | 19 911 | 20 378 | 236 | 1432 | 461 | 100 | 1016 | 5349 | 103 | 6410 | 3342 | 1668 | 8.2 |
| Cricetulus griseus | 23 884 | 24 442 | 109 | 1407 | 927 | 96 | 1170 | 7073 | 116 | 7114 | 2786 | 1516 | 6.2 |
| Mus musculus* | 43 539 | 74 158 | 1792 | 4350 | 1698 | 717 | 3443 | 20 456 | 714 | 23 226 | 9137 | 6142 | 8.3 |
| Rattus norvegicus* | 27 340 | 33 555 | 966 | 2211 | 637 | 476 | 1473 | 9153 | 411 | 10 094 | 5407 | 3177 | 9.5 |
| Spermophilus tridecemlineatus | 19 966 | 20 079 | 110 | 1437 | 429 | 83 | 937 | 5488 | 103 | 6603 | 3290 | 1547 | 7.7 |
| Primates | |||||||||||||
| Macaca fascicularis * | 17 396 | 28 955 | 233 | 1912 | 928 | 359 | 1976 | 7639 | 121 | 8186 | 2970 | 2145 | 7.4 |
| M. mulatta* | 35 536 | 69 567 | 407 | 4554 | 1694 | 653 | 3719 | 18 667 | 326 | 23 502 | 7295 | 4961 | 7.1 |
| Gorilla gorilla gorilla | 27 286 | 27 371 | 212 | 1994 | 676 | 218 | 1480 | 6701 | 151 | 9481 | 3358 | 2206 | 8.1 |
| Homo sapiens* | 68 049 | 13 5661 | 2020 | 6682 | 3480 | 3737 | 17 623 | 34 825 | 877 | 34 274 | 10 607 | 8702 | 6.4 |
| Pan troglodytes* | 20 126 | 33 326 | 296 | 2241 | 820 | 447 | 1825 | 7966 | 137 | 11 618 | 4190 | 2537 | 7.6 |
| Pongo abelii | 22 785 | 24 529 | 237 | 1818 | 580 | 229 | 1457 | 6452 | 168 | 8228 | 2879 | 2055 | 8.4 |
| Nomascus leucogenys | 19 734 | 19 837 | 141 | 1489 | 518 | 114 | 1143 | 5053 | 99 | 6893 | 2457 | 1630 | 8.2 |
| Callithrix jacchus* | 42 025 | 55 085 | 244 | 3776 | 1280 | 195 | 2867 | 15 064 | 308 | 20 159 | 6178 | 4020 | 7.3 |
| Otolemur garnettii | 19 930 | 20 156 | 99 | 1515 | 480 | 93 | 1022 | 5226 | 96 | 6801 | 3099 | 1614 | 8.0 |
| Carnivora | |||||||||||||
| Canis familiaris | 25 439 | 28 362 | 345 | 1813 | 595 | 385 | 1491 | 7040 | 170 | 9489 | 4047 | 2158 | 7.6 |
| Mustela putorius furo | 38 826 | 173 | 2017 | 984 | 137 | 2127 | 11 785 | 154 | 12 830 | 4073 | 2190 | 5.6 | |
| Neovison vison | 16 237 | 18 | 750 | 356 | 66 | 839 | 5636 | 52 | 5233 | 1507 | 768 | 4.7 | |
| Ailuropoda melanoleuca* | 21 136 | 35 743 | 247 | 2086 | 779 | 176 | 1746 | 9975 | 162 | 11 905 | 5133 | 2333 | 6.5 |
| Felis catus | 20 303 | 21 230 | 196 | 1406 | 483 | 108 | 1065 | 5831 | 107 | 6791 | 3091 | 1602 | 7.5 |
| Cetartiodactyla | |||||||||||||
| Bos mutus | 18 922 | 150 | 1377 | 453 | 123 | 931 | 4911 | 85 | 5854 | 3159 | 1527 | 8.1 | |
| Bos taurus* | 23 842 | 31 780 | 880 | 2215 | 620 | 508 | 1491 | 8171 | 317 | 9598 | 4492 | 3095 | 9.7 |
| Sus scrofa* | 26 054 | 33 962 | 645 | 2534 | 779 | 411 | 1560 | 9038 | 166 | 9463 | 4787 | 3179 | 9.4 |
| Camelus ferus | 20 028 | 67 | 1084 | 636 | 99 | 1132 | 5715 | 147 | 6257 | 2588 | 1151 | 5.7 | |
| Chiroptera | |||||||||||||
| Pteropus alecto | 19 520 | 19 548 | 97 | 1162 | 488 | 74 | 1160 | 5364 | 121 | 6774 | 2447 | 1259 | 6.4 |
| Myotis brandtii | 19 301 | 58 | 1032 | 432 | 90 | 938 | 5806 | 104 | 6427 | 2250 | 1090 | 5.6 | |
| M. davidii | 15 446 | 15 466 | 67 | 916 | 345 | 60 | 782 | 4530 | 73 | 5194 | 1816 | 983 | 6.4 |
| M. lucifugus | 20 650 | 20 899 | 143 | 1738 | 431 | 100 | 1052 | 5716 | 96 | 6782 | 2855 | 1881 | 9.0 |
| Other mammalia | |||||||||||||
| Loxodonta africana | 25 615 | 25 832 | 132 | 1744 | 556 | 128 | 1119 | 6554 | 129 | 8459 | 4835 | 1876 | 7.3 |
| Equus caballus | 22 676 | 27 841 | 272 | 1659 | 514 | 284 | 1042 | 8886 | 133 | 8701 | 3825 | 1931 | 6.9 |
| Tupaia chinensis | 20 824 | 20 851 | 85 | 1275 | 527 | 64 | 1149 | 5699 | 125 | 6701 | 3114 | 1360 | 6.5 |
| Sarcophilus harrisii | 22 388 | 22 565 | 107 | 1490 | 553 | 110 | 867 | 6368 | 102 | 7495 | 3495 | 1597 | 7.1 |
| Monodelphis domestica | 22 240 | 22 794 | 108 | 1505 | 485 | 84 | 1103 | 6398 | 106 | 7252 | 3930 | 1613 | 7.1 |
| Ornithorhynchus anatinus | 23 552 | 23 763 | 113 | 1202 | 698 | 103 | 1111 | 7229 | 95 | 7184 | 3157 | 1315 | 5.5 |
| Vertebrata (Testudines + Archosauria group) | |||||||||||||
| Anas platyrhynchos * | 16 377 | 31 879 | 139 | 1360 | 893 | 123 | 1269 | 10 542 | 148 | 9829 | 3316 | 1499 | 4.7 |
| Meleagris gallopavo | 16 537 | 16 991 | 114 | 892 | 413 | 75 | 673 | 5622 | 83 | 5377 | 2073 | 1006 | 5.9 |
| Gallus gallus* | 17 623 | 23 800 | 440 | 1640 | 581 | 278 | 1231 | 6403 | 147 | 7077 | 3282 | 2080 | 8.7 |
| Ficedula albicollis | 15 922 | 16 148 | 64 | 985 | 390 | 57 | 778 | 4669 | 81 | 5208 | 2021 | 1049 | 6.5 |
| Taeniopygia guttata | 18 141 | 19 724 | 85 | 716 | 432 | 77 | 972 | 6749 | 72 | 6197 | 2211 | 801 | 4.1 |
| Chelonia mydas | 19 066 | 71 | 880 | 478 | 71 | 794 | 6031 | 97 | 6384 | 2194 | 951 | 5.0 | |
| Pelodiscus sinensis | 20 784 | 126 | 1271 | 492 | 64 | 798 | 6724 | 92 | 6703 | 2683 | 1397 | 6.7 | |
| Other vertebrata | |||||||||||||
| Petromyzon marinus | 13 160 | 54 | 522 | 255 | 66 | 669 | 3945 | 34 | 3797 | 1239 | 576 | 4.4 | |
| Latimeria chalumnae | 23 429 | 23 513 | 75 | 1270 | 593 | 87 | 993 | 8117 | 116 | 7349 | 2905 | 1345 | 5.7 |
| Anolis carolinensis | 19 109 | 19 562 | 81 | 1238 | 510 | 288 | 936 | 5727 | 89 | 6435 | 2478 | 1319 | 6.7 |
| Invertebrate | |||||||||||||
| Chordata (Tunicata) | |||||||||||||
| Oikopleura dioica* | 17 050 | 29 057 | 15 | 1864 | 1177 | 116 | 1493 | 10 785 | 92 | 8060 | 2540 | 1879 | 6.5 |
| Ciona intestinalis | 17 308 | 18 639 | 28 | 1507 | 601 | 95 | 738 | 6231 | 47 | 5014 | 1812 | 1535 | 8.2 |
| C. savignyi | 20 004 | 20 117 | 45 | 822 | 359 | 52 | 697 | 7792 | 55 | 5685 | 2535 | 867 | 4.3 |
| Ecdysozoa (Arachnida) | |||||||||||||
| Tetranychus urticae | 18 082 | 18 243 | 12 | 1891 | 685 | 95 | 701 | 5986 | 59 | 3615 | 2219 | 1903 | 10.4 |
| Ixodes ricinus | 16 199 | 1 | 3554 | 819 | 136 | 848 | 3528 | 97 | 3649 | 1246 | 3555 | 21.9 | |
| I. scapularis | 20 473 | 21 162 | 21 | 1943 | 762 | 103 | 1433 | 5706 | 95 | 5905 | 1681 | 1964 | 9.3 |
| Rhipicephalus pulchellus | 11 205 | 1 | 1620 | 459 | 60 | 962 | 2380 | 49 | 3424 | 1231 | 1621 | 14.5 | |
| Ecdysozoa (Insecta) | |||||||||||||
| Drosophila mojavensis | 14 525 | 15 086 | 21 | 2049 | 347 | 95 | 832 | 4302 | 74 | 4241 | 1700 | 2070 | 13.7 |
| D. virilis | 14 456 | 14 928 | 34 | 1941 | 354 | 84 | 770 | 4308 | 57 | 4323 | 1709 | 1975 | 13.2 |
| D. erecta | 15 116 | 44 | 2220 | 362 | 71 | 873 | 3918 | 59 | 4366 | 1721 | 2264 | 15.0 | |
| D. grimshawi | 14 754 | 14 798 | 31 | 1861 | 362 | 62 | 773 | 4336 | 79 | 4195 | 1698 | 1892 | 12.8 |
| D. ananassae | 14 968 | 15 298 | 28 | 2139 | 349 | 64 | 791 | 4243 | 67 | 4503 | 1793 | 2167 | 14.2 |
| D. melanogaster* | 20 120 | 39 951 | 254 | 4761 | 923 | 269 | 2127 | 10 613 | 191 | 12 101 | 4659 | 5015 | 12.6 |
| D. persimilis | 16 754 | 16 861 | 28 | 2106 | 420 | 77 | 930 | 4846 | 68 | 5076 | 1701 | 2134 | 12.7 |
| D. pseudoobscura pseudoobscura | 17 047 | 48 | 2316 | 416 | 77 | 939 | 4632 | 71 | 5126 | 1950 | 2364 | 13.9 | |
| D. sechellia | 16 134 | 16 361 | 37 | 2250 | 410 | 71 | 936 | 4464 | 54 | 4765 | 1734 | 2287 | 14.0 |
| D. simulans | 15 354 | 19 057 | 57 | 2372 | 436 | 100 | 1028 | 5483 | 56 | 5374 | 2165 | 2429 | 12.7 |
| D. willistoni | 15 447 | 15 564 | 25 | 1875 | 355 | 77 | 815 | 4808 | 60 | 4434 | 1722 | 1900 | 12.2 |
| D. yakuba | 17 257 | 41 | 2521 | 392 | 77 | 1006 | 4752 | 59 | 5024 | 1798 | 2562 | 14.8 | |
| Megaselia scalaris | 11 463 | 11 503 | 10 | 773 | 417 | 31 | 449 | 4947 | 24 | 2402 | 639 | 783 | 6.8 |
| Anopheles darlingi | 10 447 | 11 686 | 8 | 793 | 272 | 93 | 758 | 3687 | 54 | 3971 | 1162 | 801 | 6.9 |
| A. gambiae* | 13 072 | 19 384 | 50 | 2610 | 410 | 87 | 1104 | 6027 | 57 | 4834 | 1869 | 2660 | 13.7 |
| Aedes aegypti | 16 654 | 17 683 | 54 | 2367 | 469 | 182 | 961 | 5052 | 56 | 4873 | 2008 | 2421 | 13.7 |
| Culex quinquefasciatus | 18 703 | 19 062 | 25 | 2345 | 534 | 128 | 1104 | 5751 | 73 | 5501 | 1866 | 2370 | 12.4 |
| Dendroctonus ponderosae | 23 650 | 14 | 1992 | 549 | 106 | 1153 | 8928 | 96 | 6087 | 2502 | 2006 | 8.5 | |
| Tribolium castaneum | 16 502 | 17 074 | 26 | 1717 | 423 | 66 | 846 | 5830 | 50 | 4196 | 2109 | 1743 | 10.2 |
| Apis mellifera | 10 910 | 12 299 | 65 | 757 | 267 | 81 | 390 | 4440 | 45 | 3293 | 1714 | 822 | 6.7 |
| Camponotus floridanus | 14 787 | 14 801 | 15 | 662 | 329 | 45 | 744 | 5533 | 65 | 4078 | 1346 | 677 | 4.6 |
| Acromyrmex echinatior | 13 962 | 13 970 | 17 | 592 | 327 | 36 | 847 | 5226 | 62 | 4219 | 1253 | 609 | 4.4 |
| Atta cephalotes | 18 079 | 18 113 | 16 | 753 | 597 | 99 | 1094 | 6579 | 75 | 4715 | 1559 | 769 | 4.2 |
| Solenopsis invicta | 14 193 | 14 359 | 26 | 636 | 437 | 100 | 748 | 5413 | 31 | 3508 | 1120 | 662 | 4.6 |
| Harpegnathos saltator | 15 029 | 15 042 | 17 | 739 | 329 | 46 | 696 | 5484 | 60 | 4223 | 1299 | 756 | 5.0 |
| Nasonia vitripennis | 17 040 | 17 289 | 14 | 1545 | 305 | 65 | 701 | 6951 | 55 | 4883 | 1423 | 1559 | 9.0 |
| Bombyx mori | 14 767 | 17 915 | 125 | 1773 | 379 | 108 | 806 | 6293 | 54 | 4580 | 1681 | 1898 | 10.6 |
| Danaus plexippus | 16 253 | 16 358 | 34 | 1486 | 441 | 95 | 808 | 5657 | 66 | 4528 | 1493 | 1520 | 9.3 |
| Pararge aegeria | 15 104 | 12 | 428 | 561 | 75 | 850 | 5983 | 14 | 3763 | 503 | 440 | 2.9 | |
| Rhodnius prolixus | 15 180 | 16 639 | 44 | 1420 | 537 | 41 | 562 | 6782 | 62 | 3769 | 1473 | 1464 | 8.8 |
| Acyrthosiphon pisum | 35 809 | 35 211 | 24 | 1834 | 814 | 102 | 1736 | 15 209 | 66 | 8622 | 1736 | 1858 | 5.3 |
| Pediculus humanus subsp. corporis | 10 847 | 11 | 513 | 257 | 37 | 349 | 4294 | 40 | 3174 | 1193 | 524 | 4.8 | |
| Ecdysozoa (Nematoda) | |||||||||||||
| Ascaris suum* | 9213 | 18 539 | 39 | 1223 | 577 | 107 | 1302 | 5894 | 75 | 4965 | 2437 | 1262 | 6.8 |
| Pristionchus pacificus | 29 079 | 29 319 | 14 | 3027 | 1038 | 75 | 1368 | 9699 | 94 | 7157 | 3263 | 3041 | 10.4 |
| Caenorhabditis brenneri | 29 982 | 30 712 | 21 | 3602 | 896 | 76 | 1134 | 10 314 | 88 | 7338 | 4255 | 3623 | 11.8 |
| C. briggsae | 21 751 | 21 914 | 30 | 2734 | 655 | 119 | 874 | 6435 | 106 | 5178 | 3540 | 2764 | 12.6 |
| C. elegans | 26 173 | 26 447 | 182 | 3573 | 856 | 163 | 1065 | 7156 | 107 | 6190 | 4401 | 3755 | 14.2 |
| C. japonica | 35 063 | 35 069 | 14 | 2665 | 998 | 95 | 2061 | 12 267 | 123 | 9112 | 3234 | 2679 | 7.6 |
| C. remanei | 31 252 | 32 133 | 21 | 3859 | 1117 | 84 | 1282 | 10 352 | 93 | 7199 | 4981 | 3880 | 12.1 |
| Haemonchus contortus | 18 580 | 3 | 2181 | 558 | 83 | 1016 | 5826 | 77 | 4679 | 2261 | 2184 | 11.8 | |
| Brugia malayi* | 1643 | 11 561 | 10 | 668 | 347 | 37 | 579 | 4540 | 44 | 3139 | 908 | 678 | 5.9 |
| Loa loa | 15 319 | 15 356 | 11 | 784 | 588 | 46 | 750 | 5774 | 49 | 3749 | 1387 | 795 | 5.2 |
| Wuchereria bancrofti | 19 298 | 19 525 | 18 | 716 | 677 | 129 | 870 | 8254 | 39 | 4504 | 1205 | 734 | 3.8 |
| Trichinella spiralis | 16 041 | 16 278 | 17 | 935 | 770 | 73 | 980 | 5389 | 71 | 3433 | 1234 | 952 | 5.8 |
| Ecdysozoa (Arthropoda) | |||||||||||||
| Daphnia pulex | 30 137 | 30 988 | 22 | 2827 | 892 | 333 | 1432 | 11 433 | 88 | 8315 | 2130 | 2849 | 9.2 |
| Strigamia maritima | 14 972 | 15 011 | 19 | 1118 | 428 | 48 | 726 | 5331 | 68 | 3635 | 1835 | 1137 | 7.6 |
| Lophotrochozoa | |||||||||||||
| Helobdella robusta | 23 328 | 23 379 | 19 | 1170 | 671 | 59 | 924 | 9385 | 145 | 5866 | 2226 | 1189 | 5.1 |
| Capitella teleta | 31 207 | 22 | 2183 | 907 | 76 | 1263 | 10 827 | 106 | 7765 | 3917 | 2205 | 7.1 | |
| Crassostrea gigas | 25 982 | 26 850 | 26 | 1904 | 633 | 85 | 814 | 10 178 | 140 | 7045 | 2912 | 1930 | 7.2 |
| Lottia gigantea | 23 721 | 34 | 1683 | 588 | 48 | 659 | 9382 | 76 | 5530 | 2734 | 1717 | 7.2 | |
| Platyhelminthes | |||||||||||||
| Echinococcus granulosus | 11 124 | 0 | 614 | 375 | 381 | 656 | 2855 | 40 | 3518 | 1260 | 614 | 5.5 | |
| E. multilocularis | 10 572 | 0 | 591 | 326 | 91 | 656 | 2878 | 48 | 3532 | 1239 | 591 | 5.6 | |
| Clonorchis sinensis | 13 606 | 13 880 | 6 | 562 | 349 | 55 | 990 | 4294 | 89 | 5074 | 1234 | 568 | 4.1 |
| Schistosoma japonicum | 16 236 | 17 | 1767 | 607 | 70 | 853 | 6086 | 36 | 3511 | 1357 | 1784 | 11.0 | |
| S. mansoni | 11 723 | 12 836 | 9 | 605 | 427 | 203 | 505 | 4491 | 60 | 3740 | 1242 | 614 | 4.8 |
| Other Invertebrates | |||||||||||||
| Amphimedon queenslandica | 29 741 | 29 816 | 6 | 1490 | 893 | 65 | 1246 | 11 722 | 73 | 7333 | 2685 | 1496 | 5.0 |
| Nematostella vectensis | 24 435 | 25 035 | 61 | 1135 | 586 | 58 | 1005 | 8651 | 72 | 6385 | 3293 | 1196 | 4.8 |
| Strongylocentrotus purpuratus | 28 567 | 29 560 | 46 | 2198 | 737 | 94 | 1101 | 9895 | 145 | 8580 | 4026 | 2244 | 7.6 |
| Trichoplax adhaerens | 11 520 | 11 590 | 7 | 482 | 213 | 36 | 489 | 5013 | 44 | 2502 | 1776 | 489 | 4.2 |
| Branchiostoma floridae | 28 544 | 29 237 | 37 | 2227 | 710 | 152 | 1146 | 8799 | 140 | 7800 | 3826 | 2264 | 7.7 |
Notes : Data of other protein subcellular locations are summarized in Supplementary Table 1 . HLS: highly likely secreted; LS: likely secreted; Mito: mitochondrial; mem: membrane; non-mem: non-membrane; Cyt: cytoplasm (or cytosol); Nuc: nuclear; Secr: secretome. Species labeled with * has more (or less) 5000 protein entries than its reference proteome.
It should be noted that the distribution data of protein subcellular locations in Table 2 and Supplementary Table S1 were based on all available protein entries for each species in the database, which were different from a complete or reference proteome in some species. Several species had more redundant proteins in the dataset. For example, human reference proteome contained 68 049 proteins while a total of 135 661 human proteins were retrieved and used for analysis ( Table 2 ). Thus, the proportions of each subcellular proteome might be slightly different for some species when a reference proteome was used. The two largest compartments having a large proportion of proteins were cytoplasm and nucleus ( Table 2 ). The proteins located in cytoplasm, not including cytoskeleton proteins, accounted for 21–43% (average 31%), and the proteins located in nucleus accounted for 20–37% (average 30%) of total proteins in these species. Approximately 3–19% (average 12%) of total proteins are predicted to be plasma membrane proteins, and 3–9% of proteins (average 5.6%) are predicted to be located in mitochondria. We noticed that 15.7% of human proteins are located in mitochondria. This number is much higher than the proportions in other species. This might be due to relatively a large number (∼7000) of curated human mitochondrial proteins in the dataset. Also, the prediction sensitivity for mitochondrial proteins was relatively low (∼42.5%) ( Table 1 ), thereby likely underestimating the proportions of mitochondrial proteins in animal species reported here.
Classical secreted proteins from a species, i.e. secretome, can be relatively accurately predicted. Combining curated secreted proteins and predicted highly likely secreted proteins (at least 3 positives out of 4 predictors) as a secretome, our method for a secretome prediction reached a MCC of 0.89 with 93.5% in sensitivity and 96.0% in specificity ( Table 1 ). The proportions of secretomes vary from 2.9% to 21.9% with an average of 8.1% in animal species . Pararge aegeria , the Speckled Wood butterfly, had the smallest secretome of 440 proteins (2.9%), and Homo sapiens (human) has the largest secretome of 8702 proteins with 2020 proteins curated as secreted. However, human protein dataset contained a large proportion of redundant entries. After mapping to the human reference proteome, a total of 4969 secreted proteins (∼7.3%) were identified (see next section, Table 3 ). After excluding species having a large number (>5000 proteins) of duplicated protein entries (species labeled with * in Table 2 ) and using human secreted proteins mapped to human reference proteome, we plotted the secretome size and proteome size of remaining 103 species ( Figure 1 ). Overall there is a good correlation between the proteome size (X) and the secretome size (Y) with a correlation coefficient of 0.658 ( Y = 289.9 + 0.066X). However, clearly the secretome size is not only determined by its proteome size in a species. There are variations among different species. For example, secretomes in mammals had a range of 4.7–9.7% (average 7.3%), while the proportions of secretomes in insecta were more variable from 2.9 to 15% (average 9.8%), with Drosophila species had an average of 13.5% secretome ( Table 2 ). We also noticed that among five species in Caenorhabditis , four exhibited a secretome accounting >11% of its proteome ( Table 2 ). Caenorhabditis is a genus of nematodes that live in bacteria-rich environments like compost piles, decaying dead animals and rotting fruit. Their large secretomes may be related to their lifestyle for digesting complex biomolecules. Recently Suh and Hutter identified 3484 putative secreted proteins C. elegans , which were retrieved from WormBase ( 34 ). Interestingly, their retrieved numbers for potential secreted proteins and trasmembrane proteins (5458) in C. elegans closely coincide with our predictions (3755 secreted proteins and 5548 transmembrane proteins).
Relationship between the predicted secretome size and the proteome size in metazoa.
The secretome size and the proportion of secretome relative to their reference proteomes in different primates
| . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . |
|---|---|---|---|---|---|---|---|---|---|
| Secretome | 4969 | 3204 | 2198 | 1460 | 2848 | 1617 | 1604 | 1852 | 1923 |
| Secretome (%) | 7.3 | 7.6 | 8.1 | 8.4 | 8.0 | 8.2 | 8.0 | 9.2 | 8.4 |
| . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . |
|---|---|---|---|---|---|---|---|---|---|
| Secretome | 4969 | 3204 | 2198 | 1460 | 2848 | 1617 | 1604 | 1852 | 1923 |
| Secretome (%) | 7.3 | 7.6 | 8.1 | 8.4 | 8.0 | 8.2 | 8.0 | 9.2 | 8.4 |
Note : The reference proteome size can be found in Table 2 . Hsap: Homo sapiens; Cjar: Callithrix jacchus; Ggor: Gorilla gorilla gorilla; Mfas: Macaca fascicularis; Mmul: Macaca lulatta; Nleu: Nomascus leucogenys; Ogar: Otolemur garnettii; Ptro: Pan troglodytes; Pabe: Pongo abelii .
The secretome size and the proportion of secretome relative to their reference proteomes in different primates
| . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . |
|---|---|---|---|---|---|---|---|---|---|
| Secretome | 4969 | 3204 | 2198 | 1460 | 2848 | 1617 | 1604 | 1852 | 1923 |
| Secretome (%) | 7.3 | 7.6 | 8.1 | 8.4 | 8.0 | 8.2 | 8.0 | 9.2 | 8.4 |
| . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . |
|---|---|---|---|---|---|---|---|---|---|
| Secretome | 4969 | 3204 | 2198 | 1460 | 2848 | 1617 | 1604 | 1852 | 1923 |
| Secretome (%) | 7.3 | 7.6 | 8.1 | 8.4 | 8.0 | 8.2 | 8.0 | 9.2 | 8.4 |
Note : The reference proteome size can be found in Table 2 . Hsap: Homo sapiens; Cjar: Callithrix jacchus; Ggor: Gorilla gorilla gorilla; Mfas: Macaca fascicularis; Mmul: Macaca lulatta; Nleu: Nomascus leucogenys; Ogar: Otolemur garnettii; Ptro: Pan troglodytes; Pabe: Pongo abelii .
Comparative analysis of secretomes in primates
Completely analysing the secretomes of all species mentioned above ( Table 2 ) is beyond the scope of this work. Here we selected the secretomes of nine primates for comparative analysis ( Table 3 ). As there are some redundant entries in the dataset, we mapped the identified secreted proteins to the reference or complete proteomes that are compiled by UniProtKB ( http://www.uniprot.org/taxonomy/complete-proteomes ). Among the nine primate species, the proportions of secretomes remained unchanged in three of them and others showed a slight increase, for example, the proportion of human secretome increased from 6.4% in the whole collection to 7.3% in the complete proteome set ( Tables 2 and 3 ). Among the nine primate species, human has the largest proteome consisting of 68 049 proteins and the largest secretome size consisting of 4,969 proteins ( Table 3 ). The large proteome size in human is mainly due to intensive collection of proteins generated by alternative splicing of protein coding genes ( 35 , 36 ). We also noted that Macaca mulatta has a much larger, nearly doubled, proteome and secretome size than M. fascicularis has ( Table 3 ). Whether such a large difference in these two closely related species is caused by the extensive genome segment duplications in M. mulatta ( 37 ) needs to be further examined.
To provide an overview of the functionalities of primate secreted proteins, we categorized the predicted secreted proteins into protein families using the rpsBLAST tool to search the Pfam database with a cutoff E-value of 1e−10. The secretomes of primates can be classified into a total of 841 unique protein families. The summary of the Pfam analysis with 28 families having 17 or more entries in a family in human is shown in Table 4 . A complete list can be found in Supplementary Table S2 . The top 10 highly encoded secreted protein families in primates were Trypsin, Immunoglobulin V-set domain, Serpin (serine protease inhibitor), Small cytokines (intecrine/chemokine), wnt family, von Willebrand factor type A domain, Immunoglobulin I-set domain, Fibrinogen beta and gamma chains, CUB domain and C1q domain. There are both variations in the Pfam categories and the number of entries in each Pfam among different primates. The significance of these secreted proteins in primate development and evolution certainly needs to be further investigated.
Comparison of protein families in primate secretomes
| Pfam ID . | Pfam Name . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . | Pfam description . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total . | . | 2586 . | 2222 . | 1573 . | 992 . | 1907 . | 1128 . | 1187 . | 1300 . | 1349 . | . |
| pfam00089 | Trypsin | 148 | 100 | 94 | 54 | 92 | 58 | 76 | 78 | 77 | Trypsin |
| pfam07686 | V-set | 72 | 100 | 61 | 93 | 77 | 21 | 49 | 13 | 106 | Immunoglobulin V-set domain |
| pfam00079 | Serpin | 60 | 30 | 23 | 22 | 25 | 16 | 20 | 20 | 23 | Serpin (serine protease inhibitor) |
| pfam00048 | IL8 | 42 | 34 | 35 | 28 | 38 | 34 | 23 | 34 | 33 | Small cytokines (intecrine/chemokine) |
| pfam00110 | wnt | 42 | 36 | 25 | 16 | 26 | 21 | 19 | 22 | 20 | wnt family |
| pfam00092 | VWA | 39 | 51 | 29 | 12 | 24 | 17 | 20 | 23 | 18 | von Willebrand factor type A domain |
| pfam07679 | I-set | 37 | 28 | 16 | 13 | 21 | 14 | 9 | 22 | 12 | Immunoglobulin I-set domain |
| pfam00147 | Fibrinogen_C | 32 | 37 | 25 | 14 | 24 | 21 | 19 | 19 | 22 | Fibrinogen beta and gamma chains |
| pfam00431 | CUB | 32 | 23 | 12 | 4 | 20 | 6 | 8 | 9 | 9 | CUB domain |
| pfam00386 | C1q | 30 | 39 | 24 | 12 | 22 | 18 | 27 | 25 | 17 | C1q domain |
| pfam00019 | TGF_beta | 25 | 30 | 29 | 18 | 27 | 20 | 25 | 23 | 23 | Transforming growth factor beta like domain |
| pfam00754 | F5_F8_type_C | 25 | 9 | 4 | 4 | 4 | 5 | 4 | 5 | 8 | F5/8 type C domain |
| pfam01403 | Sema | 25 | 20 | 8 | 7 | 11 | 10 | 3 | 7 | 6 | Sema domain |
| pfam00413 | Peptidase_M10 | 24 | 17 | 21 | 14 | 18 | 15 | 13 | 15 | 12 | Matrixin |
| pfam00059 | Lectin_C | 23 | 38 | 27 | 16 | 21 | 18 | 15 | 16 | 18 | Lectin C-type domain |
| pfam05986 | ADAM_spacer1 | 23 | 31 | 18 | 9 | 19 | 13 | 13 | 16 | 17 | ADAM-TS Spacer 1 |
| pfam00151 | Lipase | 22 | 10 | 10 | 7 | 8 | 9 | 7 | 7 | 7 | Lipase |
| pfam00061 | Lipocalin | 19 | 25 | 22 | 8 | 14 | 6 | 18 | 10 | 8 | Lipocalin/cytosolic fatty-acid binding |
| pfam00167 | FGF | 19 | 16 | 14 | 4 | 10 | 7 | 12 | 14 | 14 | Fibroblast growth factor |
| pfam00193 | Xlink | 19 | 17 | 10 | 5 | 14 | 6 | 7 | 8 | 8 | Extracellular link domain |
| pfam02931 | Neur_chan_LBD | 19 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Neurotransmitter-gated ion-channel ligand |
| pfam03024 | Folate_rec | 19 | 6 | 4 | 3 | 3 | 3 | 4 | 4 | 5 | Folate receptor family |
| pfam00530 | SRCR | 18 | 6 | 3 | 0 | 3 | 3 | 4 | 4 | 4 | Scavenger receptor cysteine-rich domain |
| pfam00055 | Laminin_N | 17 | 26 | 14 | 3 | 22 | 9 | 10 | 11 | 9 | Laminin N-terminal (Domain VI) |
| pfam00143 | Interferon | 17 | 11 | 14 | 8 | 16 | 11 | 10 | 13 | 13 | Interferon alpha/beta domain |
| pfam00246 | Peptidase_M14 | 17 | 18 | 14 | 8 | 15 | 12 | 9 | 14 | 12 | Zinc carboxypeptidase |
| pfam07546 | EMI | 17 | 10 | 8 | 3 | 7 | 4 | 7 | 7 | 3 | EMI domain |
| pfam13895 | Ig_2 | 17 | 5 | 2 | 0 | 6 | 3 | 1 | 8 | 1 | Immunoglobulin domain |
| Pfam ID . | Pfam Name . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . | Pfam description . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total . | . | 2586 . | 2222 . | 1573 . | 992 . | 1907 . | 1128 . | 1187 . | 1300 . | 1349 . | . |
| pfam00089 | Trypsin | 148 | 100 | 94 | 54 | 92 | 58 | 76 | 78 | 77 | Trypsin |
| pfam07686 | V-set | 72 | 100 | 61 | 93 | 77 | 21 | 49 | 13 | 106 | Immunoglobulin V-set domain |
| pfam00079 | Serpin | 60 | 30 | 23 | 22 | 25 | 16 | 20 | 20 | 23 | Serpin (serine protease inhibitor) |
| pfam00048 | IL8 | 42 | 34 | 35 | 28 | 38 | 34 | 23 | 34 | 33 | Small cytokines (intecrine/chemokine) |
| pfam00110 | wnt | 42 | 36 | 25 | 16 | 26 | 21 | 19 | 22 | 20 | wnt family |
| pfam00092 | VWA | 39 | 51 | 29 | 12 | 24 | 17 | 20 | 23 | 18 | von Willebrand factor type A domain |
| pfam07679 | I-set | 37 | 28 | 16 | 13 | 21 | 14 | 9 | 22 | 12 | Immunoglobulin I-set domain |
| pfam00147 | Fibrinogen_C | 32 | 37 | 25 | 14 | 24 | 21 | 19 | 19 | 22 | Fibrinogen beta and gamma chains |
| pfam00431 | CUB | 32 | 23 | 12 | 4 | 20 | 6 | 8 | 9 | 9 | CUB domain |
| pfam00386 | C1q | 30 | 39 | 24 | 12 | 22 | 18 | 27 | 25 | 17 | C1q domain |
| pfam00019 | TGF_beta | 25 | 30 | 29 | 18 | 27 | 20 | 25 | 23 | 23 | Transforming growth factor beta like domain |
| pfam00754 | F5_F8_type_C | 25 | 9 | 4 | 4 | 4 | 5 | 4 | 5 | 8 | F5/8 type C domain |
| pfam01403 | Sema | 25 | 20 | 8 | 7 | 11 | 10 | 3 | 7 | 6 | Sema domain |
| pfam00413 | Peptidase_M10 | 24 | 17 | 21 | 14 | 18 | 15 | 13 | 15 | 12 | Matrixin |
| pfam00059 | Lectin_C | 23 | 38 | 27 | 16 | 21 | 18 | 15 | 16 | 18 | Lectin C-type domain |
| pfam05986 | ADAM_spacer1 | 23 | 31 | 18 | 9 | 19 | 13 | 13 | 16 | 17 | ADAM-TS Spacer 1 |
| pfam00151 | Lipase | 22 | 10 | 10 | 7 | 8 | 9 | 7 | 7 | 7 | Lipase |
| pfam00061 | Lipocalin | 19 | 25 | 22 | 8 | 14 | 6 | 18 | 10 | 8 | Lipocalin/cytosolic fatty-acid binding |
| pfam00167 | FGF | 19 | 16 | 14 | 4 | 10 | 7 | 12 | 14 | 14 | Fibroblast growth factor |
| pfam00193 | Xlink | 19 | 17 | 10 | 5 | 14 | 6 | 7 | 8 | 8 | Extracellular link domain |
| pfam02931 | Neur_chan_LBD | 19 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Neurotransmitter-gated ion-channel ligand |
| pfam03024 | Folate_rec | 19 | 6 | 4 | 3 | 3 | 3 | 4 | 4 | 5 | Folate receptor family |
| pfam00530 | SRCR | 18 | 6 | 3 | 0 | 3 | 3 | 4 | 4 | 4 | Scavenger receptor cysteine-rich domain |
| pfam00055 | Laminin_N | 17 | 26 | 14 | 3 | 22 | 9 | 10 | 11 | 9 | Laminin N-terminal (Domain VI) |
| pfam00143 | Interferon | 17 | 11 | 14 | 8 | 16 | 11 | 10 | 13 | 13 | Interferon alpha/beta domain |
| pfam00246 | Peptidase_M14 | 17 | 18 | 14 | 8 | 15 | 12 | 9 | 14 | 12 | Zinc carboxypeptidase |
| pfam07546 | EMI | 17 | 10 | 8 | 3 | 7 | 4 | 7 | 7 | 3 | EMI domain |
| pfam13895 | Ig_2 | 17 | 5 | 2 | 0 | 6 | 3 | 1 | 8 | 1 | Immunoglobulin domain |
Note : A complete list is shown as Supplementary Table 2 . The species full names can be found in the note of Table 3 .
Comparison of protein families in primate secretomes
| Pfam ID . | Pfam Name . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . | Pfam description . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total . | . | 2586 . | 2222 . | 1573 . | 992 . | 1907 . | 1128 . | 1187 . | 1300 . | 1349 . | . |
| pfam00089 | Trypsin | 148 | 100 | 94 | 54 | 92 | 58 | 76 | 78 | 77 | Trypsin |
| pfam07686 | V-set | 72 | 100 | 61 | 93 | 77 | 21 | 49 | 13 | 106 | Immunoglobulin V-set domain |
| pfam00079 | Serpin | 60 | 30 | 23 | 22 | 25 | 16 | 20 | 20 | 23 | Serpin (serine protease inhibitor) |
| pfam00048 | IL8 | 42 | 34 | 35 | 28 | 38 | 34 | 23 | 34 | 33 | Small cytokines (intecrine/chemokine) |
| pfam00110 | wnt | 42 | 36 | 25 | 16 | 26 | 21 | 19 | 22 | 20 | wnt family |
| pfam00092 | VWA | 39 | 51 | 29 | 12 | 24 | 17 | 20 | 23 | 18 | von Willebrand factor type A domain |
| pfam07679 | I-set | 37 | 28 | 16 | 13 | 21 | 14 | 9 | 22 | 12 | Immunoglobulin I-set domain |
| pfam00147 | Fibrinogen_C | 32 | 37 | 25 | 14 | 24 | 21 | 19 | 19 | 22 | Fibrinogen beta and gamma chains |
| pfam00431 | CUB | 32 | 23 | 12 | 4 | 20 | 6 | 8 | 9 | 9 | CUB domain |
| pfam00386 | C1q | 30 | 39 | 24 | 12 | 22 | 18 | 27 | 25 | 17 | C1q domain |
| pfam00019 | TGF_beta | 25 | 30 | 29 | 18 | 27 | 20 | 25 | 23 | 23 | Transforming growth factor beta like domain |
| pfam00754 | F5_F8_type_C | 25 | 9 | 4 | 4 | 4 | 5 | 4 | 5 | 8 | F5/8 type C domain |
| pfam01403 | Sema | 25 | 20 | 8 | 7 | 11 | 10 | 3 | 7 | 6 | Sema domain |
| pfam00413 | Peptidase_M10 | 24 | 17 | 21 | 14 | 18 | 15 | 13 | 15 | 12 | Matrixin |
| pfam00059 | Lectin_C | 23 | 38 | 27 | 16 | 21 | 18 | 15 | 16 | 18 | Lectin C-type domain |
| pfam05986 | ADAM_spacer1 | 23 | 31 | 18 | 9 | 19 | 13 | 13 | 16 | 17 | ADAM-TS Spacer 1 |
| pfam00151 | Lipase | 22 | 10 | 10 | 7 | 8 | 9 | 7 | 7 | 7 | Lipase |
| pfam00061 | Lipocalin | 19 | 25 | 22 | 8 | 14 | 6 | 18 | 10 | 8 | Lipocalin/cytosolic fatty-acid binding |
| pfam00167 | FGF | 19 | 16 | 14 | 4 | 10 | 7 | 12 | 14 | 14 | Fibroblast growth factor |
| pfam00193 | Xlink | 19 | 17 | 10 | 5 | 14 | 6 | 7 | 8 | 8 | Extracellular link domain |
| pfam02931 | Neur_chan_LBD | 19 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Neurotransmitter-gated ion-channel ligand |
| pfam03024 | Folate_rec | 19 | 6 | 4 | 3 | 3 | 3 | 4 | 4 | 5 | Folate receptor family |
| pfam00530 | SRCR | 18 | 6 | 3 | 0 | 3 | 3 | 4 | 4 | 4 | Scavenger receptor cysteine-rich domain |
| pfam00055 | Laminin_N | 17 | 26 | 14 | 3 | 22 | 9 | 10 | 11 | 9 | Laminin N-terminal (Domain VI) |
| pfam00143 | Interferon | 17 | 11 | 14 | 8 | 16 | 11 | 10 | 13 | 13 | Interferon alpha/beta domain |
| pfam00246 | Peptidase_M14 | 17 | 18 | 14 | 8 | 15 | 12 | 9 | 14 | 12 | Zinc carboxypeptidase |
| pfam07546 | EMI | 17 | 10 | 8 | 3 | 7 | 4 | 7 | 7 | 3 | EMI domain |
| pfam13895 | Ig_2 | 17 | 5 | 2 | 0 | 6 | 3 | 1 | 8 | 1 | Immunoglobulin domain |
| Pfam ID . | Pfam Name . | Hsap . | Cjar . | Ggor . | Mfas . | Mmul . | Nleu . | Ogar . | Ptro . | Pabe . | Pfam description . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total . | . | 2586 . | 2222 . | 1573 . | 992 . | 1907 . | 1128 . | 1187 . | 1300 . | 1349 . | . |
| pfam00089 | Trypsin | 148 | 100 | 94 | 54 | 92 | 58 | 76 | 78 | 77 | Trypsin |
| pfam07686 | V-set | 72 | 100 | 61 | 93 | 77 | 21 | 49 | 13 | 106 | Immunoglobulin V-set domain |
| pfam00079 | Serpin | 60 | 30 | 23 | 22 | 25 | 16 | 20 | 20 | 23 | Serpin (serine protease inhibitor) |
| pfam00048 | IL8 | 42 | 34 | 35 | 28 | 38 | 34 | 23 | 34 | 33 | Small cytokines (intecrine/chemokine) |
| pfam00110 | wnt | 42 | 36 | 25 | 16 | 26 | 21 | 19 | 22 | 20 | wnt family |
| pfam00092 | VWA | 39 | 51 | 29 | 12 | 24 | 17 | 20 | 23 | 18 | von Willebrand factor type A domain |
| pfam07679 | I-set | 37 | 28 | 16 | 13 | 21 | 14 | 9 | 22 | 12 | Immunoglobulin I-set domain |
| pfam00147 | Fibrinogen_C | 32 | 37 | 25 | 14 | 24 | 21 | 19 | 19 | 22 | Fibrinogen beta and gamma chains |
| pfam00431 | CUB | 32 | 23 | 12 | 4 | 20 | 6 | 8 | 9 | 9 | CUB domain |
| pfam00386 | C1q | 30 | 39 | 24 | 12 | 22 | 18 | 27 | 25 | 17 | C1q domain |
| pfam00019 | TGF_beta | 25 | 30 | 29 | 18 | 27 | 20 | 25 | 23 | 23 | Transforming growth factor beta like domain |
| pfam00754 | F5_F8_type_C | 25 | 9 | 4 | 4 | 4 | 5 | 4 | 5 | 8 | F5/8 type C domain |
| pfam01403 | Sema | 25 | 20 | 8 | 7 | 11 | 10 | 3 | 7 | 6 | Sema domain |
| pfam00413 | Peptidase_M10 | 24 | 17 | 21 | 14 | 18 | 15 | 13 | 15 | 12 | Matrixin |
| pfam00059 | Lectin_C | 23 | 38 | 27 | 16 | 21 | 18 | 15 | 16 | 18 | Lectin C-type domain |
| pfam05986 | ADAM_spacer1 | 23 | 31 | 18 | 9 | 19 | 13 | 13 | 16 | 17 | ADAM-TS Spacer 1 |
| pfam00151 | Lipase | 22 | 10 | 10 | 7 | 8 | 9 | 7 | 7 | 7 | Lipase |
| pfam00061 | Lipocalin | 19 | 25 | 22 | 8 | 14 | 6 | 18 | 10 | 8 | Lipocalin/cytosolic fatty-acid binding |
| pfam00167 | FGF | 19 | 16 | 14 | 4 | 10 | 7 | 12 | 14 | 14 | Fibroblast growth factor |
| pfam00193 | Xlink | 19 | 17 | 10 | 5 | 14 | 6 | 7 | 8 | 8 | Extracellular link domain |
| pfam02931 | Neur_chan_LBD | 19 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Neurotransmitter-gated ion-channel ligand |
| pfam03024 | Folate_rec | 19 | 6 | 4 | 3 | 3 | 3 | 4 | 4 | 5 | Folate receptor family |
| pfam00530 | SRCR | 18 | 6 | 3 | 0 | 3 | 3 | 4 | 4 | 4 | Scavenger receptor cysteine-rich domain |
| pfam00055 | Laminin_N | 17 | 26 | 14 | 3 | 22 | 9 | 10 | 11 | 9 | Laminin N-terminal (Domain VI) |
| pfam00143 | Interferon | 17 | 11 | 14 | 8 | 16 | 11 | 10 | 13 | 13 | Interferon alpha/beta domain |
| pfam00246 | Peptidase_M14 | 17 | 18 | 14 | 8 | 15 | 12 | 9 | 14 | 12 | Zinc carboxypeptidase |
| pfam07546 | EMI | 17 | 10 | 8 | 3 | 7 | 4 | 7 | 7 | 3 | EMI domain |
| pfam13895 | Ig_2 | 17 | 5 | 2 | 0 | 6 | 3 | 1 | 8 | 1 | Immunoglobulin domain |
Note : A complete list is shown as Supplementary Table 2 . The species full names can be found in the note of Table 3 .
We further performed Gene Ontology (GO) analysis with the human secretome by searching the UniProtKB/Swiss-Prot dataset using BLASTP with a cutoff E-value of 1e−10. GO information was retrieved from UniProt ID mapping data ( http://www.uniprot.org/downloads ) and analysed using GO SlimViewer with generic GO terms ( 38 ). Among 4969 human secreted proteins, 4,512 entries had at least one GO mapping. As the proteins in the dataset are predicted to be secreted, thus, only GO biological process and molecular function classification is further analysed ( Figure 2 ; Supplementary Table S3 ). Secreted proteins in humans are involved in 67 biological processes with a total of 25,887 GO IDs. The top five processes include anatomical structure development (13.8%), signal transduction (9.7%), immune system process (7.5%), response to stress (6.3%), and cell differentiation (5.8%) ( Figure 2 a). Molecular function analysis revealed human secreted proteins had 39 types of molecular functions with a total of 3,059 GO IDs. The top five main molecular functions include ion binding (28.5%), peptidase activity (11.8%), signal transducer activity (9.9%), enzyme regulator activity (7.5%) and oxidoreductase activity (5.9%) ( Figure 2 b). GO analysis and functional protein family domain analysis are consistent in showing these proteins play important roles in signal transduction, immune system, regulation of human structure development and many other biological processes.
Gene Ontology classification of the human secreted protein distribution in ( a ) biological process and ( b ) molecular function ontology.
Discussion
The work described here represents our efforts to computationally predict the subcellular locations for all human and animal proteins, with a focus on secretomes. In addition, for the secretomes, we further classified them as curated, predicted to be highly likely secreted, likely secreted, and weakly likely secreted protein subsets. This refinement of classifications of secreted proteins and other subcellular locations is expected to greatly facilitate comparative analysis of subcellular proteomes in different species. Human secretome research is an active research subject due to its importance in human health and medicine, such as the human secretome atlas initiative with a goal for identifying potential biomarkers and therapeutic targets in the secretome that can be traced back in accessible human body fluids ( 12 ). For example, recently the human secreted enzyme Notum was found to inhibit the Wnt signaling pathway through removal of a lipid that is linked to the Wnt proteins and that is required for activation of Wnt receptor proteins ( 39 , 40 ). Analysis of the secretome can yield valuable data leading to an understanding of the intricate interaction between different tissues as it relates to the coordination of physiology in multicellular organisms. An example is found in the interaction between muscles and bones ( 41 ). Many muscle specific growth factors, in the myosecretome, have been shown to have effects on bone repair and remodeling. Myostatin, a myocyte derived growth factor that inhibits muscle growth and thus acting as a break on uncontrolled growth, also has effects on suppression of bone marrow-derived stem cells and cartilage formation ( 41 ). In this study, we compared secretomes in different primates, and revealed that the highly enriched families including Trypsin, Immunoglobulin V-set domain, Serpin (serine protease inhibitor), Small cytokines (intecrine/chemokine) and wnt family, etc. Further we analysed the molecular functions and biological processes of the human secretome. Our analysis revealed the secreted proteins in humans play important roles in human structure development, immune systems, and response to stress, etc.
In this work, the secretome identification was limited to classical secreted proteins, i.e. signal peptide containing proteins, and curated secreted proteins that may include both classical and leadless-secreted proteins (LSP). SecretomeP was a tool implemented for predicting these LSPs in bacteria and mammals ( http://www.cbs.dtu.dk/services/SecretomeP/ ). Because the accuracy of this tool for predicting animal LSPs is not evaluated, we did not include this tool in our data processing. Thus we would like to request the research community to submit metazoan protein subcellular locations, particularly LSPs, with experimental evidence traceable from literature to the database. The information provided in the database, the easy to download feature, and BLAST tool to allow users to search all protein data or the secretome data will provide useful supports to researcher working in these subjects. Researchers working with a new protein sequence can predict protein subcellular locations using the tools we have used in this work or other available tools that were summarized by Meinken and Min ( 32 ) and Caccia et al. ( 42 ).
The LOCATE database was developed for the human and mouse protein subcellular locations using multiple sources of information including literature data and computational prediction ( 17 ). However, the limit of the database was only for human and mouse proteins and the database has not been updated since 2009. Recently a new database named COMPARTMENTS was developed for seven model organisms including yeast, Arabidopsis, human, mouse, rat, fruit fly and Caenorhabditis elegans ( http://compartments.jensenlab.org ) ( 43 ). Our database contains protein data from all available metazoan species, with 121 species or subspecies having a complete proteome, including these model organisms. For plant and fungal protein data, we have specifically developed the plant secretome and subcellular proteome knowledgebase (PlantSecKB) ( 30 ) and the fungal secretome and subcellular proteome knowledgebase (FunSecKB and FunSecKB2) ( 3 , 4 ). The COMPARTMENTS database was implemented by integrating information from UniProtKB, STRING, GO annotations from respective model organism databases, text mining, as well as prediction information using WoLF PSORT and YLoc-HighRes methods. In comparing with our database, both used the annotation information from UniProtKB and WoLF PSORT was the common tool used for prediction information. However, some other tools are used in our database development including TargetP, SignalP, Phobius, TMHMM and PS-Scan. In contrast, the COMPARTMENTS database used YLoc-HighRes method and also STRING, GO annotations. And also the COMPARTMENTS database has developed an automatically updated web resource to update from the major eukaryotic model organisms. Our database remained static for the predicted information and will be updated periodically for manually curated data based on the literature. Thus LOCATE, COMPARTMETNS and MetazSecKB may complement each other as each of them had specific features derived from different sources or prediction tools. Therefore, we recommend researchers to cross search these databases for proteins from model organisms. However, we noticed that these databases used different identifiers for protein entries, thus the data may not be compared directly. We anticipate the MetazSecKB, along with our published fungal secretome and subcellular proteome knowledgebase (FunSecKB2) ( 4 ) and the newly developed protist secretome and subcellular proteome knowledgebase (ProtSecKB) ( http://proteomics.ysu.edu/secretomes/protist/index.php ), will serve the community valuable resources for proteome-wide comparative analysis and for investigating protein–protein interactions of host and fungal or protist pathogens.
Acknowledgements
We thank Dr. Feng Yu for his assistance in maintaining the server. The work is supported by the College of Science, Technology, Engineering, and Mathematics Dean’s reassigned time for research to X.J.M.
Funding
The work is funded by University Research Council (URC), Youngstown State University (YSU). J.M. was supported with a graduate research assistantship by the Center for Applied Chemical Biology, YSU. Funding for open access charge: the College of Graduate Studies and Research, YSU.
Conflict of interest . None declared.
References
Author notes
Present address: John Meinken, Center for Health Informatics, University of Cincinnati, Cincinnati, OH 45267-0840, USA
Citation details: Meinken,J., Walker,G., Cooper,C.R., et al . MetazSecKB: the human and animal secretome and subcellular proteome knowledgebase. Database (2015) Vol. 2015: article ID bav077; doi:10.1093/database/bav077