-
PDF
- Split View
-
Views
-
Cite
Cite
Tuan-Dung Le, Phuong D Nguyen, Dmitry Korkin, Thanh Thieu, PHILM2Web: A high-throughput database of macromolecular host–pathogen interactions on the Web, Database, Volume 2022, 2022, baac042, https://doi.org/10.1093/database/baac042
Close - Share Icon Share
Abstract
During infection, the pathogen’s entry into the host organism, breaching the host immune defense, spread and multiplication are frequently mediated by multiple interactions between the host and pathogen proteins. Systematic studying of host–pathogen interactions (HPIs) is a challenging task for both experimental and computational approaches and is critically dependent on the previously obtained knowledge about these interactions found in the biomedical literature. While several HPI databases exist that manually filter HPI protein–protein interactions from the generic databases and curated experimental interactomic studies, no comprehensive database on HPIs obtained from the biomedical literature is currently available. Here, we introduce a high-throughput literature-mining platform for extracting HPI data that includes the most comprehensive to date collection of HPIs obtained from the PubMed abstracts. Our HPI data portal, PHILM2Web (Pathogen–Host Interactions by Literature Mining on the Web), integrates an automatically generated database of interactions extracted by PHILM, our high-precision HPI literature-mining algorithm. Currently, the database contains 23 581 generic HPIs between 157 host and 403 pathogen organisms from 11 609 abstracts. The interactions were obtained from processing 608 972 PubMed abstracts, each containing mentions of at least one host and one pathogen organisms. In response to the coronavirus disease 2019 (COVID-19) pandemic, we also utilized PHILM to process 25 796 PubMed abstracts obtained by the same query as the COVID-19 Open Research Dataset. This COVID-19 processing batch resulted in 257 HPIs between 19 host and 31 pathogen organisms from 167 abstracts. The access to the entire HPI dataset is available via a searchable PHILM2Web interface; scientists can also download the entire database in bulk for offline processing.
Database URL: http://philm2web.live
Introduction
Infections are complex biological processes that are common among a variety of microbial pathogens, such as viruses, bacteria, fungi, protozoa, multicellular parasites and even proteins, (4, 25, 51) targeting host organisms from virtually all kingdoms of life. Infectious diseases dominated World Health Organization’s list of threats to global health (78) and have an adverse economic impact, costing billions of dollars every year (60). Human infections are also the largest part of the neglected diseases, a group of tropical diseases that are spread among the poorest segment of the world’s population (28, 29, 48). The 2019 novel coronavirus [causing coronavirus disease 2019 (COVID-19)] exemplified the devastation of a highly infectious disease spreading throughout the world via modern human mobility, resulting in more than 600 000 deaths in the USA (12) and more than 4 million deaths worldwide (79). Knowledge about animal infections also plays an important role in human disease discovery and prevention: many discovered infectious diseases of wild and domesticated animals pose a significant threat to human health (15, 30, 69). The pathogen’s strategy to enter host’s organism and breach its immune defenses often involves interactions between the host and pathogen macromolecules, including proteins, peptides, RNAs and DNAs (24, 35, 62). Understanding the molecular mechanisms of host–pathogen interactions (HPIs) is a challenging task for both experimental and computational approaches and is critically dependent on the previous knowledge about these interactions (23, 61, 66, 68). In addition, important conclusions about such interactions can be drawn from the studies of other interactions between the related host and pathogen organisms, since the molecular mechanisms underlying related infectious diseases are often common (20). Recently, there have been several approaches to gather large datasets of HPIs, either by heuristic filtering from existing protein–protein interaction (PPI) databases (6, 10, 11, 18, 37, 46, 53, 67, 80) or by manual curation of HPIs from biomedical literature, primarily for selected hosts or pathogens (3, 7, 9, 17, 63, 81). However, an automated approach that accurately and comprehensively mines the HPI data by integrating heterogeneous sources is yet to be built. One of the principal data sources currently unexplored by the HPI-mining approaches is PubMed, a database of the peer-reviewed biomedical literature, which includes more than 32 million abstracts of research papers and books. This amount of data makes it infeasible to comprehensively detect HPI-relevant abstracts and annotate the HPI manually, even with an expert-based search of the PubMed database. Therefore, there exists a need for a high-throughput system that not only mines HPI data quickly, but also facilitates a platform to navigate the mined information at scale. In this work, we assembled a high-precision, automated system that mined a comprehensive HPI database from PubMed abstracts. The rest of this paper is organized as follows: Related Work section discusses existing HPI databases and related text mining methods; Methods section presents our literature mining system and large scale information extraction from PubMed; Results section shows our database and its comparison against other popular databases; Discussion section analyzes characteristics of our work; and Conclusion section summarizes our contribution.
Related Work
Curated Databases
During the last decade, a handful of resources that manually collected HPI data have emerged and can be categorized into four groups: (G1) targeting specific hosts or pathogens, (G2) targeting pathogen families, (G3) targeting host families and (G4) heterogeneous host and pathogen families. Group G1 includes HCVpro (41) for hepatitis C virus, HIV-1 Human Interaction (2), Proteopathogen (73) for Candida albicans and HoPaCI-db (7) for Pseudomonas aeruginosa and Coxiella burnetii. Group G2 includes Viruses.STRING (17), VirHostNet (32) and VirusMINT (13) for virus pathogenicity; PIG/PATRIC (76) for all types of bacteria; InnateDB (9) for immune response of humans, mices and bovines to microbial infection and PHIDIAS (81) for virulence factors of 100 pathogens. Group G3 includes PHISTO (22) for all pathogen types interacting with human, MorCVD (63) for cardiovascular diseases and BioGrid (65) for interactions from Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens. Group G4 includes PHI-base (71) and HPIDB (3, 40), the largest HPI data source that integrated information from other databases. In rare databases that did include automated processing, text mining was often an insignificant, ad hoc component. For example, Viruses.STRING (17) had an entity detection module for virus species and proteins (16, 55) based on dictionary matching, PHISTO (22) had a custom text-mining module to extract names of the experimental methods. The work closest to ours is the HPIDB database, which contains manually curated HPIs at the macromolecular level. While HPIDB focuses on expert manual curation of task-specific HP–PPIs, our work focuses on large-scale automatic mining of HP–PPIs from scientific literature.
Literature Mining
Literature mining (or text mining) of PPIs can be categorized into two groups: (i) host–pathogen interspecies interactions and (ii) generic PPIs. A review of computational system biology showed that literature mining of HPI was underdeveloped (21). For example, pathogen-specific text mining of HPI was only focused on Brucella (36). Simpler systems used information retrieval search engine to find associations between human diseases, genes, proteins and drugs (47). Besides text mining, another group of approaches characterized the interaction structures by applying interaction network (49, 56, 72), interspecies homology (8), sequencing information (5) and microarray analysis (45). On the other hand, literature mining of generic PPIs has been well-studied, exemplified by a number of research community initiatives, such as BioCreAtIve and BioNLP, and ongoing meetings and workshops (34, 38, 39, 44, 52, 59). Recent methodologies in this track made use of deep neural network architectures such as Convolutional Neural Networks (CNN) (14, 57), Long Short-Term Memory (LSTM) networks (1, 82), multi-head attention (1, 84) and transformers (75). Our work integrates the previous results of PHILM on general HPI (70) with recent interactions relating to COVID-19 from PubMed. Our text-mining method was based on pattern matching on the dependency graphs of parsed sentences, an approach proven to generate higher-precision results than both the statistical learning and deep learning counterparts (70, 83).
Methods
Literature Mining of HPIs
We utilized an updated version of the original PHILM system (70) to mine information about HPI from PubMed abstracts. PHILM used link grammars (31) to analyze dependency structures of text sentences and then extracted HPI information using pattern matching. A HPI extraction system is similar to a general PPI extraction system, with additional challenges including (i) correct association of the organism for each protein, (ii) ensuring that the extracted interaction is an inter- and not intraspecies interaction and (iii) combining the information about an HPI across multiple sentences. In this update, we replaced NLProt (50) gene normalization functionality by bridging BANNER (43) with SR4GN (77).
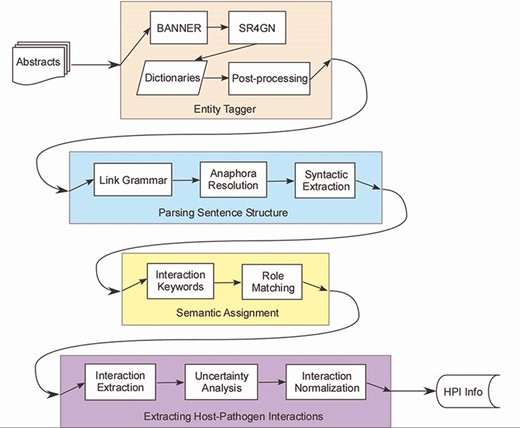
PHILM consisted of four phases (Figure 1): (i) entity tagging, where proteins/genes and organism names were identified and linked according to species–gene relationship; (ii) parsing sentences structure, where input text was parsed into dependency structures that allowed resolution of anaphora to pronouns, and splitting a complex sentence into simple sentences; (iii) semantic assignment, where HPI roles of components of a simple sentence were determined and (iv) extraction of HPI information, where both host and pathogenic parties of an interaction were localized, together with interaction keywords, sentence index, uncertainty analysis of the interaction and normalizing the interaction across sentences.
Host–pathogen literature mining system. PHILM consisted of four phases: (i) entity tagging, where proteins/genes and organism names were identified and linked according to species-gene relationship; (ii) parsing sentence structure, where input text was parsed into dependency structures that allowed resolution of anaphors and splitting a complex sentence into simple sentences; (iii) semantic assignment, where HPI roles of components of a simple sentence were determined and (iv) extraction of HPI information, where both host and pathogen parties of an interaction were localized, normalized and analyzed for uncertainty.
Entity Tagging
This phase identified proteins/genes names and names of organisms associated with the proteins/genes. PHILM was so sensitive to species association that it was crucial that the parent organism of a protein (which can be either a host or a pathogen) was correctly identified. This phase consisted of four modules: protein/gene tagging using BANNER, species association using SR4GN, heuristic host/pathogen dictionary matching and postprocessing.
BANNER
BANNER used conditional random field (42) together with a dictionary of 334 000 one-syllabus names to identify gene/protein names in biomedical text.
SR4GN
SR4GN used a species name dictionary combined with heuristic rules to detect species mentioned in biomedical text. After that, species names were linked to gene/protein names detected by BANNER by heuristic rules.
Dictionary matching
Organisms found by SR4GN were scanned against our host–pathogen dictionary to find their roles. To support mining COVID-19 information, our dictionary included species of the coronavirus genus as pathogen organisms. This module also grouped multiple mentions of the same protein/gene into a protein/gene entity with a unique UniProt accession number (6). Likewise, multiple mentions of the same organisms were grouped into an organism entity with a unique National Center for Biotechnology Information (NCBI) Taxonomy ID (26).
Postprocessing
This module used the phrasal structure generated by link grammar to (i) infer host/pathogen information not included in the dictionary and (ii) re-associate a protein/gene to a grammar-supported organism. In the first round, it searched for generic keywords (e.g. ‘host’, ‘pathogen’, ‘pathogenic’, ‘pathogenesis’, etc.), in each phrase that contained unidentified organism names. In the second round, the modules searched for co-existence of a protein/gene and an organism in a phrase that satisfies one of following two patterns and then overwrote the organism association suggested by SR4GN: Pattern 1: Organism name + protein name (e.g. ‘Arabidopsis RIN4 protein’) and Pattern 2: Protein name + preposition + organism name (e.g. ‘RXLX of human’).
Parsing Sentence Structure
This phase leveraged the grammatical structure of a sentence to assist with information extraction. It consisted of four modules: link grammar, three-layer entity framework, anaphora resolution and syntactic extraction.
Link grammar
Link grammar (64) relied on dependency rules to link pairs of related words. PHILM used link grammar implementation of AbiWord (http://www.abisource.com/projects/link-grammar/) that incorporated the original link grammar with an expansion to biomedical sublanguage, BioLG (58).
Three-layer entity framework
To support entity linking and normalization, PHILM implemented a hierarchy consisting of three layers that connects textual entities (in middle layer with text sentences) down to real entities (in bottom layer with UniProt and NCBI Taxonomy identification numbers) and up to link grammar nodes (in top layer with link grammar parses). Any change in host/pathogen role of an organism or protein–organism association automatically propagated to related entities via the three-layer connections.
Anaphora resolution
This module linked entities (protein/gene/organism) with respective anaphoric pronouns using Hobbs’ algorithm (33). It helped with consolidating HPI information across multiple sentences.
Syntactic extraction
This module split a complex sentence into simple sentences with four components: Subject (S) + Verb (V) + Object (O) + Modifying phrase of verb (M). The algorithm traversed the linkage structure of the complex sentence and extracted tuples of four connected link types: S link (connects a subject to a verb), RS link (connects a verb to a subject), O link (connects a verb to an object) and MV link (connects a verb to a modifying phrase).
Semantic Assignment
This phase assigned HPI-related, semantic roles to components of a simple sentence. It consisted of two modules: interaction keyword tagging and role type matching.
Interaction keyword tagging
This module identified interaction keyword at stemming level. Stems were derived from both WordNet (27) lexical database and our manually curated dictionary.
Role type matching
This module assigned a role for each syntactic component (i.e. subject, verb, object and modifying phrase) of a simple sentence. An elementary role signified that the component only contained a single host entity, a single pathogen entity or an interaction keyword. A partial role meant that the component contained two types of entities. A complete role meant that the component contained all three types of entities.
Interaction Extraction and Normalization
This phase was the end-point of PHILM that extracted and validated elements of identified host-pathogen interactions. It consisted of three modules: interaction extraction, uncertainty analysis and interaction normalization.
Interaction extraction
This module first grouped syntactic components so that each group jointly contained complete information about an HPI (i.e. host + pathogen + interaction keyword entities). After that, each group was matched against appropriate interaction patterns to extract HPI entities. For example, a pattern ‘S<E> V<E> O<E> = P<S> I<V> H<O>’ indicated that if three components of a simple sentence were both elementary, then the sentence might contain (i) a pathogen entity in its subject; (ii) an interaction keyword in its verb and (iii) a host entity in its object. PHILM used seven templates that scanned through all syntactic components of a simple sentence: subject, verb, object and modifying phrase.
Uncertainty analysis
This module scanned the sentence against negation keywords (e.g. ‘does not’ and ‘cannot’) and uncertainty keywords (e.g. ‘possibly’ and ‘may’). A negation/uncertainty keyword was in effect if there was a link connecting the keyword with any syntactic component of the simple sentence.
Interaction normalization
This module first collapsed duplicate entities that linked to the same real entity using UniProt and NCBI Taxonomy identification numbers. Then, it collapsed duplicate HPIs having the same quadruple of host/pathogen proteins/genes and organisms. Furthermore, uncertainty evidence across multiple sentences describing the same HPI were aggregated to become a unified uncertainty flag.
Large-scale Mining from PubMed
Collecting PubMed abstracts that potentially contained general HPIs
We run two customized queries against the PubMed database. The first query searched for the presence of at least one host organism in the abstracts and it returned 5 008 750 PubMed IDs. The second query searched for the presence of at least one pathogen organism in the abstracts and it returned 1 459 547 PubMed IDs. Computing set intersection on these two sets of PubMed IDs gave us 608 972 abstracts that contained at least both a host and a pathogen organism. We recorded those abstracts as potentially containing general HPI information.
Collecting PubMed abstracts that potentially contained COVID-19 HPIs
We run the same query used by the COVID-19 Open Research Dataset (74) on PubMed. The query retrieved 25 796 abstracts containing species of the coronavirus genus, including Novel Coronavirus (2019-nCoV), Severe Acute Respiratory Syndrome-associated Coronavirus (SARS-CoV), and Middle East Respiratory Syndrome Coronavirus (MERS-CoV). We recorded those abstracts as potentially containing COVID-19 HPI information.
High-throughput HPI mining
We run PHILM on our college’s high-performance computing cluster. The system run on 140 CPUs over 6 days to completely process 608 972 general HPI relevant abstracts and 25 796 COVID-19 relevant abstracts. Regarding general HPI information, the system extracted 23 581 HPI interactions between 157 host and 403 pathogen organisms from 11 609 relevant abstracts. Regarding COVID-19 HPI information, the system extracted 257 interactions between 19 host and 31 pathogen organisms from 167 relevant abstracts. All found HPI information was transferred to PHILM2Web for community benefits.
PHILM2Web web interface
We employed a low-latency, searchable web interface (https://github.com/vividvilla/csvtotable) for easy investigation and analysis of the large number of HPIs extracted from the literature. The interface allows browsing though all interactions, instantaneous filtering interactions by keywords and downloading the entire interaction database for offline analysis. Search keywords are matched against both Pubmed ID, host organisms/proteins/genes, pathogen organisms/proteins/genes and interaction keywords to facilitate flexible search. HPI database for PHILM2Web was extracted from PubMed abstracts and deposited automatically using our high-precision biomedical literature mining system developed specifically to handle the HPI information. PHILM2Web currently contains 23 581 general HPIs and 257 COVID-19-related interactions extracted from 32 million PubMed abstracts.
Results
Database
Our HPI database contains 23 581 generic HPIs and 257 COVID-19-related interactions from 11 609 relevant PubMed abstracts. Hereinafter, we denote the generic set of HPI interactions as PhilmHPI and the COVID-19-related set of interactions as PhilmCOVID. To gauge coverage of the HPI database, we compare it against two popular databases: HPIDB (3, 40) and IntAct (53). For coverage on HPI in general, we compare PhilmHPI against HPIDB, the largest manually curated database on macromolecular HPIs. Since HPIDB does not contain a section for COVID-19, we compare PhilmCOVID against the COVID-19 section of IntAct, the largest manually curated database on generic PPIs.
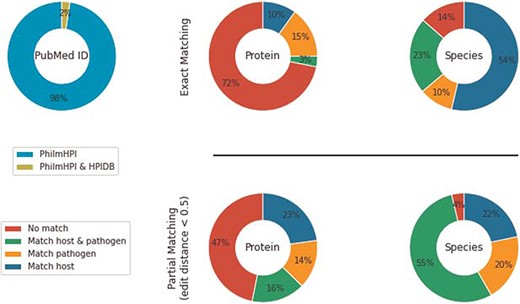
HPIDB contains 69 787 curated interactions from 4985 PubMed IDs. Among those, 280 PubMed IDs overlap with PhilmHPI. The overlap is <2.5% of total number of abstracts mined in PhilmHPI (Figure 2). We further analyze 2046 interaction pairs found in the 280 overlapping PubMed IDs. More specifically, we take one interaction from PhilmHPI and one interaction from HPIDB originating from the same Pubmed ID and then we compare their host proteins, host species, pathogen proteins and pathogen species (Figure 2).
Under exact matching, two proteins (or species) are considered matched if their IDs are the same or there is at least one exact string match among the synonyms of their names. Synonyms of a protein name are aggregated from three sources: (i) from HPIDB entry aliases derived from its respective source databases, (ii) if the HPIDB entry has a UniProt ID, then we retrieve UniProt’s recommended names, alternative names and submitted names and (3) similar to retrieving synonyms from UniProt, we also retrieve official symbol, official full name and ‘as known as’ sections from NCBI Gene ID. Results show that 10% of interactions in PhilmHPI do not match anything in HPIDB. We also observe that host/pathogen species pairs match much better than host/pathogen protein pairs. A probable reason is that species names are less diverse than protein names, and human is the dominant common host species in both databases.
We observe that while HPIDB contains standardized names taken from source databases such as NCBI and UniProt, our PhilmHPI contains article-specific names extracted from the article text. For example, abstract 15328338 abbreviates human FVT-1 gene as hFVT-1 and mouse FVT-1 gene as mFVT-1. As a result, PHILM identifies hFVT-1 and mFVT-1 as interactants. However, HPIDB links these interactants to gene IDs 2531 and 70 750, both having the same alias name FVT1. Exact matching fails because of the article-specific prefixes ‘h’ and ‘m’, together with the extra dash ‘-’ in the gene names. We present common mismatch scenarios in Table 1.
To alleviate this naming diversity issue, we also analyze databases overlap using partial matching. Like exact matching, two proteins/species are considered matched if their IDs are the same. However, when IDs are unavailable, we compute names similarity as string edit distance. We use Natural Language Toolkit (NLTK) implementation of edit distance with |$substitution\_cost = 2$| and then normalized the result by the summation of lengths of both names. Two proteins/species names are considered partially matched if the normalized edit distance score is less than an user-specified threshold. We empirically used 0.5 as the partial matching threshold after observing a large number of reasonably similar names retrieved by this threshold.
Mismatch names of the same proteins/genes between PhilmHPI versus HPIDB and PhilmCOVID versus IntActCOVID.
| PubMed ID . | PhilmHPI . | HPIDB . | Edit distance . |
|---|---|---|---|
| 15328338 | hFVT-1 | FVT1 | 0.2 |
| mFVT-1 | FVT1 | 0.2 | |
| 9405152 | Importin-beta | Importin-90 | 0.25 |
| 12198176 | E6 protein | Protein E6 | 0.3 |
| 21900157 | CLE/C14orf166 protein | C14orf166 | 0.4 |
| PubMed ID | PhilmCOVID | IntActCOVID | Edit distance |
| 21411533 | hACE2 | ACE2 | 0.11 |
| 18448518 | EF1alpha | EEF1AL | 0.29 |
| 30209168 | N protein | Nucleocapsid protein | 0.38 |
| 20861307 | Small envelope protein (E) | E protein | 0.49 |
| PubMed ID . | PhilmHPI . | HPIDB . | Edit distance . |
|---|---|---|---|
| 15328338 | hFVT-1 | FVT1 | 0.2 |
| mFVT-1 | FVT1 | 0.2 | |
| 9405152 | Importin-beta | Importin-90 | 0.25 |
| 12198176 | E6 protein | Protein E6 | 0.3 |
| 21900157 | CLE/C14orf166 protein | C14orf166 | 0.4 |
| PubMed ID | PhilmCOVID | IntActCOVID | Edit distance |
| 21411533 | hACE2 | ACE2 | 0.11 |
| 18448518 | EF1alpha | EEF1AL | 0.29 |
| 30209168 | N protein | Nucleocapsid protein | 0.38 |
| 20861307 | Small envelope protein (E) | E protein | 0.49 |
Mismatch names of the same proteins/genes between PhilmHPI versus HPIDB and PhilmCOVID versus IntActCOVID.
| PubMed ID . | PhilmHPI . | HPIDB . | Edit distance . |
|---|---|---|---|
| 15328338 | hFVT-1 | FVT1 | 0.2 |
| mFVT-1 | FVT1 | 0.2 | |
| 9405152 | Importin-beta | Importin-90 | 0.25 |
| 12198176 | E6 protein | Protein E6 | 0.3 |
| 21900157 | CLE/C14orf166 protein | C14orf166 | 0.4 |
| PubMed ID | PhilmCOVID | IntActCOVID | Edit distance |
| 21411533 | hACE2 | ACE2 | 0.11 |
| 18448518 | EF1alpha | EEF1AL | 0.29 |
| 30209168 | N protein | Nucleocapsid protein | 0.38 |
| 20861307 | Small envelope protein (E) | E protein | 0.49 |
| PubMed ID . | PhilmHPI . | HPIDB . | Edit distance . |
|---|---|---|---|
| 15328338 | hFVT-1 | FVT1 | 0.2 |
| mFVT-1 | FVT1 | 0.2 | |
| 9405152 | Importin-beta | Importin-90 | 0.25 |
| 12198176 | E6 protein | Protein E6 | 0.3 |
| 21900157 | CLE/C14orf166 protein | C14orf166 | 0.4 |
| PubMed ID | PhilmCOVID | IntActCOVID | Edit distance |
| 21411533 | hACE2 | ACE2 | 0.11 |
| 18448518 | EF1alpha | EEF1AL | 0.29 |
| 30209168 | N protein | Nucleocapsid protein | 0.38 |
| 20861307 | Small envelope protein (E) | E protein | 0.49 |
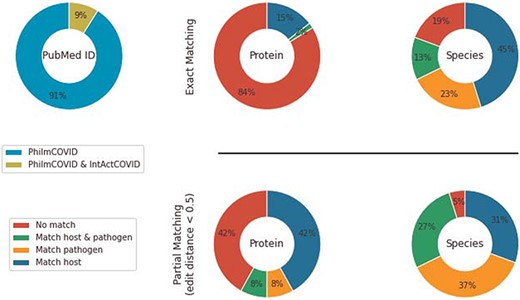
IntAct’s COVID-19 section (IntAct COVID) contains 7315 COVID-19-related, generic PPIs from 256 PubMed IDs. Among those, 15 PubMed IDs overlapped with PhilmCOVID. We used the host–pathogen species hierarchy in PHILM (70) (https://academic.oup.com/view-large/83412208) to filter HPIs from IntAct COVID. Specifically, (i) NCBI taxonomy lineage of interactant species were retrieved, (ii) species were classified into different levels in accordance with the host–pathogen species hierarchy and (iii) an organism from a higher level could only be a host of an organism from a lower level. After filtering and duplicate removal, we compared the remaining 62 interactions from IntAct COVID against PhilmCOVID using the same exact matching and partial matching strategies (Figure 3).
Web Interface
Our web interface presents HPI information in a tabular format with 12 columns for PubMed ID, host/pathogen interactants, interaction keywords, indexes of sentences where the interactions occur and confidence level (Figure 4). Each row in the table represents one HPI interaction between one host protein/gene and one pathogen protein/gene. For easy reference, we link abstract IDs to PubMed, organism/species IDs to NCBI Taxonomy (26) and protein/gene IDs to UniProt (6). The entire database can be downloaded in JSON or CSV format by clicking corresponding buttons on the top-left corner. Investigators can use the search box on the top-right corner to instantaneously filter HPI interactions on the web. To report error, an investigator clicks the red exclamation mark button on the left of the interaction row to open an error reporting form displayed at the bottom of the web page.
Evaluation
We evaluated the accuracy of our text mining system on two benchmarking sets: (1) GenericBenchmark contains 266 generic HPI interactions in 175 PubMed abstracts obtained from the original PHILM evaluation (70) and (2) CovidBenchmark contains 281 COVID-19-related interactions from 167 PubMed abstracts in PhilmCOVID. The GenericBenchmark inherits human labeling from the original PHILM evaluation (70). P.D.N., who holds a Ph.D. in biology, manually labeled interactions in CovidBenchmark. In calculating performance measures, an exact match is counted when an extracted name (i.e. protein name, gene name or organism name) either matches exactly to a corresponding human labeled name or refers to the same biological entity determined by human expert. We also report performance measures on partial matching, that is when two names having a normalized edit distance <0.5. Table 2 presents full evaluation results on individual gene/protein, individual organism, pairs of genes/proteins, pairs of organisms and complete HPI interaction.
Accuracy of PHILM2Web assessed over two manually labeled datasets: GenericBenchmark and CovidBenchmark. P: precision, R: recall, F1: F1-score
| . | . | GenericBenchmark . | CovidBenchmark . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Exact Match . | Partial Match . | Exact Match . | Partial Match . | ||||||||
| . | . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . |
| Gene/Protein | Host | 0.63 | 0.13 | 0.22 | 0.65 | 0.14 | 0.23 | 0.65 | 0.62 | 0.63 | 0.79 | 0.75 | 0.77 |
| Pathogen | 0.54 | 0.15 | 0.23 | 0.67 | 0.18 | 0.29 | 0.38 | 0.4 | 0.39 | 0.43 | 0.46 | 0.45 | |
| Pair | 0.4 | 0.08 | 0.13 | 0.46 | 0.09 | 0.15 | 0.16 | 0.14 | 0.15 | 0.17 | 0.16 | 0.16 | |
| Organism | Host | 0.67 | 0.2 | 0.31 | 0.67 | 0.2 | 0.31 | 0.78 | 0.75 | 0.77 | 0.78 | 0.75 | 0.77 |
| Pathogen | 0.58 | 0.17 | 0.26 | 0.65 | 0.19 | 0.29 | 0.54 | 0.49 | 0.52 | 0.63 | 0.58 | 0.61 | |
| Pair | 0.33 | 0.06 | 0.11 | 0.4 | 0.08 | 0.13 | 0.26 | 0.24 | 0.25 | 0.3 | 0.27 | 0.28 | |
| HPI Interaction | 0.15 | 0.03 | 0.05 | 0.21 | 0.04 | 0.07 | 0.09 | 0.08 | 0.08 | 0.11 | 0.1 | 0.1 | |
| . | . | GenericBenchmark . | CovidBenchmark . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Exact Match . | Partial Match . | Exact Match . | Partial Match . | ||||||||
| . | . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . |
| Gene/Protein | Host | 0.63 | 0.13 | 0.22 | 0.65 | 0.14 | 0.23 | 0.65 | 0.62 | 0.63 | 0.79 | 0.75 | 0.77 |
| Pathogen | 0.54 | 0.15 | 0.23 | 0.67 | 0.18 | 0.29 | 0.38 | 0.4 | 0.39 | 0.43 | 0.46 | 0.45 | |
| Pair | 0.4 | 0.08 | 0.13 | 0.46 | 0.09 | 0.15 | 0.16 | 0.14 | 0.15 | 0.17 | 0.16 | 0.16 | |
| Organism | Host | 0.67 | 0.2 | 0.31 | 0.67 | 0.2 | 0.31 | 0.78 | 0.75 | 0.77 | 0.78 | 0.75 | 0.77 |
| Pathogen | 0.58 | 0.17 | 0.26 | 0.65 | 0.19 | 0.29 | 0.54 | 0.49 | 0.52 | 0.63 | 0.58 | 0.61 | |
| Pair | 0.33 | 0.06 | 0.11 | 0.4 | 0.08 | 0.13 | 0.26 | 0.24 | 0.25 | 0.3 | 0.27 | 0.28 | |
| HPI Interaction | 0.15 | 0.03 | 0.05 | 0.21 | 0.04 | 0.07 | 0.09 | 0.08 | 0.08 | 0.11 | 0.1 | 0.1 | |
Accuracy of PHILM2Web assessed over two manually labeled datasets: GenericBenchmark and CovidBenchmark. P: precision, R: recall, F1: F1-score
| . | . | GenericBenchmark . | CovidBenchmark . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Exact Match . | Partial Match . | Exact Match . | Partial Match . | ||||||||
| . | . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . |
| Gene/Protein | Host | 0.63 | 0.13 | 0.22 | 0.65 | 0.14 | 0.23 | 0.65 | 0.62 | 0.63 | 0.79 | 0.75 | 0.77 |
| Pathogen | 0.54 | 0.15 | 0.23 | 0.67 | 0.18 | 0.29 | 0.38 | 0.4 | 0.39 | 0.43 | 0.46 | 0.45 | |
| Pair | 0.4 | 0.08 | 0.13 | 0.46 | 0.09 | 0.15 | 0.16 | 0.14 | 0.15 | 0.17 | 0.16 | 0.16 | |
| Organism | Host | 0.67 | 0.2 | 0.31 | 0.67 | 0.2 | 0.31 | 0.78 | 0.75 | 0.77 | 0.78 | 0.75 | 0.77 |
| Pathogen | 0.58 | 0.17 | 0.26 | 0.65 | 0.19 | 0.29 | 0.54 | 0.49 | 0.52 | 0.63 | 0.58 | 0.61 | |
| Pair | 0.33 | 0.06 | 0.11 | 0.4 | 0.08 | 0.13 | 0.26 | 0.24 | 0.25 | 0.3 | 0.27 | 0.28 | |
| HPI Interaction | 0.15 | 0.03 | 0.05 | 0.21 | 0.04 | 0.07 | 0.09 | 0.08 | 0.08 | 0.11 | 0.1 | 0.1 | |
| . | . | GenericBenchmark . | CovidBenchmark . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Exact Match . | Partial Match . | Exact Match . | Partial Match . | ||||||||
| . | . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . | P . | R . | F1 . |
| Gene/Protein | Host | 0.63 | 0.13 | 0.22 | 0.65 | 0.14 | 0.23 | 0.65 | 0.62 | 0.63 | 0.79 | 0.75 | 0.77 |
| Pathogen | 0.54 | 0.15 | 0.23 | 0.67 | 0.18 | 0.29 | 0.38 | 0.4 | 0.39 | 0.43 | 0.46 | 0.45 | |
| Pair | 0.4 | 0.08 | 0.13 | 0.46 | 0.09 | 0.15 | 0.16 | 0.14 | 0.15 | 0.17 | 0.16 | 0.16 | |
| Organism | Host | 0.67 | 0.2 | 0.31 | 0.67 | 0.2 | 0.31 | 0.78 | 0.75 | 0.77 | 0.78 | 0.75 | 0.77 |
| Pathogen | 0.58 | 0.17 | 0.26 | 0.65 | 0.19 | 0.29 | 0.54 | 0.49 | 0.52 | 0.63 | 0.58 | 0.61 | |
| Pair | 0.33 | 0.06 | 0.11 | 0.4 | 0.08 | 0.13 | 0.26 | 0.24 | 0.25 | 0.3 | 0.27 | 0.28 | |
| HPI Interaction | 0.15 | 0.03 | 0.05 | 0.21 | 0.04 | 0.07 | 0.09 | 0.08 | 0.08 | 0.11 | 0.1 | 0.1 | |
First, we notice that PHILM2Web has lower F1-scores and recalls on GenericBenchmark than CovidBenchmark across all measurement categories. The reason is GenericBenchmark includes multiple host and pathogen species that makes HPI information more diverse and complex. Second, partial matching performance is higher than exact matching performance on both benchmarks due to the relaxed matching requirement. Third, PHILM2Web excels in high precision, especially in GenericBenchmark where precision is about five times recall across all measurement categories. Given the high-throughout nature of PHILM2Web, higher precision is desirable to assist investigators in surfing the literature.
Case Studies
To illustrate utility of the PHILM2Web database, we present several PubMed abstracts that exist in both PHILM2Web and either HPIDB or IntAct. We notice that PHILM2Web captures verbatim entity names reported by authors of the abstracts, while HPIDB and IntAct map the names to standardized vocabularies such as NCBI (26) and UniProt (6). As a consequence, PHILM2Web is often more specific about protein/gene names than HPIDB and IntAct. We also present several abstracts that are exclusive to PHILM2Web to illustrate the coverage issue of manual curation pertaining to HPIDB and IntAct.
Case Study 1
PubMed abstract 5113910: Generic HPI found in both PHILM2Web and HPIDB. In this abstract, both PHILM2Web and HPIDB capture H. sapiens as host organism and Hepatitis C virus as pathogen organism. While PHILM2Web detects human RNA helicase as the original wording about host protein in the abstract, HPIDB captures the same host protein aligned to a longer name probable ATP-dependent RNA helicase DDX5 in UniProt (6). Regarding pathogen protein, PHILM2Web detects the specific protein NS5B as HCV RNA-dependent RNA polymerase reported in the abstract, while HPIDB shows both the specific NS5B protein and a vague genome polyprotein that comprises several subgroup proteins including RNA-dependent RNA polymerase.
Case Study 2
PubMed abstract 20484023: Generic HPI exclusive to PHILM2Web. In this abstract, PHILM2Web detects the plant host protein RPM1 in Arabidopsis thaliana and the pathogen virulence proteins AvrB in Pseudomonas syringae. Both HPIDB and IntAct have no information about this abstract.
Case Study 3
PubMed abstract 18448518: COVID-related HPI found in both PHILM2Web and IntAct. In this abstract, both PHILM2Web and IntAct capture H. sapiens as host organism and Severe acute respiratory syndrome-related coronavirus as pathogen organism. Regarding host protein, both PHILM2Web and IntAct detect the human elongation factor1-alpha protein, but PHILM2Web keeps the name EF1alpha reported in the abstract while IntAct shows a standardized name EEF1A2 from UniProt (6). Both PHILM2Web and IntAct detect N protein as pathogen protein.
Case Study 4
PubMed abstract 17581748: COVID-related HPI exclusive to PHILM2Web. In this abstract, PHILM2Web identifies H. sapiens host protein Nab and pathogen protein SARS-CoV spike (S) glycoprotein in SARS coronavirus. Both HPIDB and IntAct have no information about this abstract.
Discussion
Mechanistic understanding of HPI is important for pathogenicity and infectious disease research. Our web-enabled high-throughput database of HPIs extracted from PubMed abstracts provides an efficient tool for investigators to screen findings reported in the literature, thus avoiding unnecessary laboratory experiments and shortening the time to develop a cure. PHILM2Web database utilizes a high-precision specialized text-mining system that emphasizes on the accuracy of extracted information. In this information-overwhelming era, providing users with highly accurate HPI information helps save them from spending time and effort validating false-positive interactions. Nevertheless, our current database only captures HPI information in scientific abstracts, leaving potentially relevant information in other sections of a scientific paper unexplored. We did not extract deep aspects of a macromolecular interaction such as interaction detection method, binding type and author-provided confidence score according to IMEx standard (54). Our data are equivalent to shallow interaction information that comprises primary components of an HPI. Finally, our database only covers HPI reported in PubMed, the official source of peer-reviewed, published papers. Information from non-peer-reviewed, preprint sources such as bioRxiv, medRxiv and others also carries value but is not yet processed by our method. In the future, we anticipate to customize neural network architectures such as BERT (19) to improve retrieval precision. Having a high-precision information extraction system is important to maintain users’ trust for an automated method. We will also explore HPI information in all sections of a full paper and extract-relevant IMEx aspects of a macromolecular interaction.
Conclusion
We presented PHILM2Web, a web-based, high-throughput tool for biologist and healthcare researchers to investigate macromolecular HPIs reported in scientific literature. We focused on a high-precision literature-mining system to provide high-quality information and efficiently engage users. Comparison against other manually curated, expensive human-labored databases (HPIDB and IntAct) showed that our database not only has healthy overlap with the manual databases, but also contains a large number of HPI not included in the manual databases. Our database covers more than twice the number of PubMed IDs compared to HPIDB, and a slightly less number of COVID-19-relevant PubMed IDs compared to IntAct. To illustrate the accuracy and usefulness of the database, we validated it over two manually curated benchmarks and provided users’ case studies. We envision our contribution will accelerate research in infectious diseases, pandemic control and therapeutics.
Acknowledgement
We thank Samantha Warren and Andi Dhroso for their feedback and review of the manuscript.