-
PDF
- Split View
-
Views
-
Cite
Cite
Ashley Clayton, Mialy DeFelice, Brynn Zalmanek, Jay Hodgson, Caroline Morin, Stockard Simon, Julie A Bletz, James A Eddy, Milen Nikolov, Jineta Banerjee, Kalyan Vinnakota, Marco Marasca, Kevin J Boske, Bruce Hoff, Ljubomir Bradic, YooRi Kim, James R Goss, Robert J Allaway, Centralizing neurofibromatosis experimental tool knowledge with the NF Research Tools Database, Database, Volume 2022, 2022, baac045, https://doi.org/10.1093/database/baac045
Close - Share Icon Share
Abstract
Experimental tools and resources, such as animal models, cell lines, antibodies, genetic reagents and biobanks, are key ingredients in biomedical research. Investigators face multiple challenges when trying to understand the availability, applicability and accessibility of these tools. A major challenge is keeping up with current information about the numerous tools available for a particular research problem. A variety of disease-agnostic projects such as the Mouse Genome Informatics database and the Resource Identification Initiative curate a number of types of research tools. Here, we describe our efforts to build upon these resources to develop a disease-specific research tool resource for the neurofibromatosis (NF) research community. This resource, the NF Research Tools Database, is an open-access database that enables the exploration and discovery of information about NF type 1-relevant animal models, cell lines, antibodies, genetic reagents and biobanks. Users can search and explore tools, obtain detailed information about each tool as well as read and contribute their observations about the performance, reliability and characteristics of tools in the database. NF researchers will be able to use the NF Research Tools Database to promote, discover, share, reuse and characterize research tools, with the goal of advancing NF research.
Database URL: https://tools.nf.synapse.org/.
Introduction
Research tools and reagents, such as animal models, cell lines and antibodies, are essential to designing and executing successful biological experiments. Research tools are typically generated over years of work from many laboratories and are often developed to tackle specific scientific questions. Once developed, these tools can be useful for a variety of scientific purposes, yet many tools remain accessible to a limited number of investigators, or worse, unknown to the research community at large.
There are many challenges in identifying tools that can be beneficial to one’s research goals. First, given the fast pace of scientific research, it can be difficult to keep up with emerging tools. Second, scientists lack disease-specific venues to document and share important information about new applications, observations and changes for research tools prior to publication. Third, for those new to a particular research field or disease area, understanding the repertoire of tools available to them is a major impediment. Our experience in the neurofibromatosis (NF) field has shown that researchers often struggle to identify and locate available research tools and understand which tool is best suited for their experiment.
Databases that maintain a catalog of available tools help manage the above-mentioned challenges. Examples include foundational model system-specific databases, such as the Mouse Genome Informatics database (1), the Rat Genome Database (2), the Zebrafish Information Network (3) and the Saccharomyces Genome Database (4), and vendor catalogs that manufacture specific tools like antibodies, cell lines and genetic reagents. Recent efforts like the Resource Identification Initiative (5) integrate information for a wide variety of research tools, including cell lines via Cellosaurus (6), animal models via databases like Mutant Mouse Resource & Research Centers (7), antibodies via Antibody Registry (8) and reagents for genetic experiments (specifically, plasmids) via Addgene (9). The Resource Identification Initiative collaborates with these tool-specific authorities who assign research resource identifiers (RRIDs) to all of these research tools, enabling researchers to cite well-defined and curated records of specific tools. While these databases are critical sources of information, they are disease-agnostic and may not always provide the depth of information that researchers need to identify and apply a particular tool to their domain or disease.
We developed the NF Research Tools Database to mitigate these and other obstacles (Table 1) for the NF research community. To make the database easy to explore, we created a publicly available companion web application (tools.nf.synapse.org), accessible through the existing NF Data Portal (10). This database, currently in the prototype phase, catalogs a wide variety of NF type 1 (NF1)-relevant research tools. Specifically, we aggregated and curated metadata for NF1-relevant animal models, cell lines, genetic reagents, antibodies and biobanks. We then developed a web-based application that enables interactive searching and filtering of the database. The application also allows users to actively contribute by sharing reliability, biology, usage and other observations for each tool and suggesting new resources for inclusion. Finally, the tools are linked to the NF Data Portal (10), so, when available, data generated with or from these tools can be more easily discovered and accessed.
Challenges faced by the NF community in finding research tools, our perspective on how an NF-specific research tools database helps address these challenges (‘with an NF-specific tool database’) and our assessment of how an investigator might struggle without this database (‘without an NF-specific tool database’)
| Challenges faced by investigators . | With an NF-specific tool database . | Without an NF-specific tool database . |
|---|---|---|
| Unaware of central databases for research tools | The NF Data Portal is an established and highly visible portal in this community; by layering a tools database on top, we can make investigators aware of tools, as well as drive traffic to and awareness of the RRID tool authorities | Investigators may remain unaware of tool-specific repositories and tools |
| NF is a rare disease and has few relevant tools; central databases that catalog all tools can be overwhelming | Disease-specific tool portal has a very narrow, deep focus and does not contain tools that are not relevant for NF | Investigators must search/filter through large databases to identify tools of interest, which may be time-consuming |
| No place to deposit ‘unpublishable’ observations about tool performance | We have created a mechanism for investigators to deposit and share observations about NF tools | Observations are limited to personal communications, personal websites, etc. |
| Finding tool-associated data are difficult | This tool database is within the NF Data Portal and has direct links to data generated using the tools | Would need to search the NF Data Portal for tool-related data and may need to try multiple synonyms for the same tool |
| Organizations like Gilbert Family Foundation are attempting to expand the research community to non-NF1 researchers who have no prior knowledge of NF-related tools | An NF-focused database could lower the barrier of entry to researchers that are new to the field | New researchers may not have a good frame of reference for what is ‘NF-relevant’ vs. not |
| Central tool databases do not always have prepublication/in-development models | With our unique positioning as a data coordinating center for many NF projects and our relationships with NF funders and researchers, we are able to facilitate cataloging of tools that are in-development | Tools will not be cataloged/listed by central authorities until publication |
| Challenges faced by investigators . | With an NF-specific tool database . | Without an NF-specific tool database . |
|---|---|---|
| Unaware of central databases for research tools | The NF Data Portal is an established and highly visible portal in this community; by layering a tools database on top, we can make investigators aware of tools, as well as drive traffic to and awareness of the RRID tool authorities | Investigators may remain unaware of tool-specific repositories and tools |
| NF is a rare disease and has few relevant tools; central databases that catalog all tools can be overwhelming | Disease-specific tool portal has a very narrow, deep focus and does not contain tools that are not relevant for NF | Investigators must search/filter through large databases to identify tools of interest, which may be time-consuming |
| No place to deposit ‘unpublishable’ observations about tool performance | We have created a mechanism for investigators to deposit and share observations about NF tools | Observations are limited to personal communications, personal websites, etc. |
| Finding tool-associated data are difficult | This tool database is within the NF Data Portal and has direct links to data generated using the tools | Would need to search the NF Data Portal for tool-related data and may need to try multiple synonyms for the same tool |
| Organizations like Gilbert Family Foundation are attempting to expand the research community to non-NF1 researchers who have no prior knowledge of NF-related tools | An NF-focused database could lower the barrier of entry to researchers that are new to the field | New researchers may not have a good frame of reference for what is ‘NF-relevant’ vs. not |
| Central tool databases do not always have prepublication/in-development models | With our unique positioning as a data coordinating center for many NF projects and our relationships with NF funders and researchers, we are able to facilitate cataloging of tools that are in-development | Tools will not be cataloged/listed by central authorities until publication |
Challenges faced by the NF community in finding research tools, our perspective on how an NF-specific research tools database helps address these challenges (‘with an NF-specific tool database’) and our assessment of how an investigator might struggle without this database (‘without an NF-specific tool database’)
| Challenges faced by investigators . | With an NF-specific tool database . | Without an NF-specific tool database . |
|---|---|---|
| Unaware of central databases for research tools | The NF Data Portal is an established and highly visible portal in this community; by layering a tools database on top, we can make investigators aware of tools, as well as drive traffic to and awareness of the RRID tool authorities | Investigators may remain unaware of tool-specific repositories and tools |
| NF is a rare disease and has few relevant tools; central databases that catalog all tools can be overwhelming | Disease-specific tool portal has a very narrow, deep focus and does not contain tools that are not relevant for NF | Investigators must search/filter through large databases to identify tools of interest, which may be time-consuming |
| No place to deposit ‘unpublishable’ observations about tool performance | We have created a mechanism for investigators to deposit and share observations about NF tools | Observations are limited to personal communications, personal websites, etc. |
| Finding tool-associated data are difficult | This tool database is within the NF Data Portal and has direct links to data generated using the tools | Would need to search the NF Data Portal for tool-related data and may need to try multiple synonyms for the same tool |
| Organizations like Gilbert Family Foundation are attempting to expand the research community to non-NF1 researchers who have no prior knowledge of NF-related tools | An NF-focused database could lower the barrier of entry to researchers that are new to the field | New researchers may not have a good frame of reference for what is ‘NF-relevant’ vs. not |
| Central tool databases do not always have prepublication/in-development models | With our unique positioning as a data coordinating center for many NF projects and our relationships with NF funders and researchers, we are able to facilitate cataloging of tools that are in-development | Tools will not be cataloged/listed by central authorities until publication |
| Challenges faced by investigators . | With an NF-specific tool database . | Without an NF-specific tool database . |
|---|---|---|
| Unaware of central databases for research tools | The NF Data Portal is an established and highly visible portal in this community; by layering a tools database on top, we can make investigators aware of tools, as well as drive traffic to and awareness of the RRID tool authorities | Investigators may remain unaware of tool-specific repositories and tools |
| NF is a rare disease and has few relevant tools; central databases that catalog all tools can be overwhelming | Disease-specific tool portal has a very narrow, deep focus and does not contain tools that are not relevant for NF | Investigators must search/filter through large databases to identify tools of interest, which may be time-consuming |
| No place to deposit ‘unpublishable’ observations about tool performance | We have created a mechanism for investigators to deposit and share observations about NF tools | Observations are limited to personal communications, personal websites, etc. |
| Finding tool-associated data are difficult | This tool database is within the NF Data Portal and has direct links to data generated using the tools | Would need to search the NF Data Portal for tool-related data and may need to try multiple synonyms for the same tool |
| Organizations like Gilbert Family Foundation are attempting to expand the research community to non-NF1 researchers who have no prior knowledge of NF-related tools | An NF-focused database could lower the barrier of entry to researchers that are new to the field | New researchers may not have a good frame of reference for what is ‘NF-relevant’ vs. not |
| Central tool databases do not always have prepublication/in-development models | With our unique positioning as a data coordinating center for many NF projects and our relationships with NF funders and researchers, we are able to facilitate cataloging of tools that are in-development | Tools will not be cataloged/listed by central authorities until publication |
Materials and Methods
Initial user story development
The concept for this database was inspired by informal conversations with members of the NF1 research community, and an initial proof of concept database was created using a sampling of research tools. Building on this initial database, anecdotal evidence from community discussions and internal brainstorming from experience in NF1 research, we developed a set of user stories (Supplementary Table S1), for example, ‘as a clinician-scientist, I want to find out what NF1, NF2, SPRED1, TP53 or SUZ12 mutations a disease model has so that I know if the model has the genetic mutation profile that I am interested in studying’. We then prioritized the user stories and designed a database and web application for high-priority user stories.
Data model development
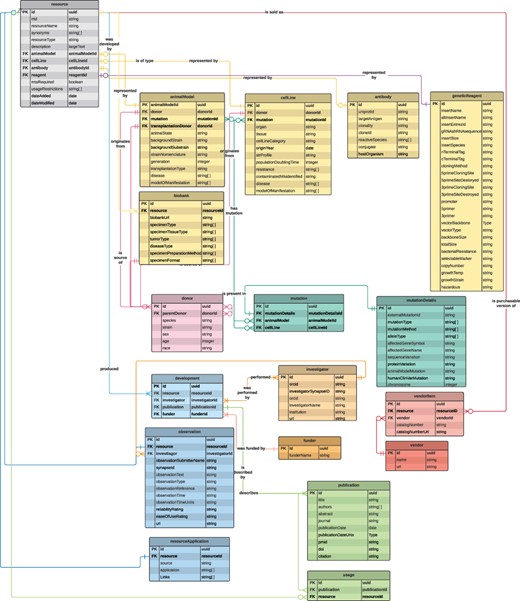
We designed a data model (Figure 1) for the database by evaluating several sources of information for cell lines (Cellosaurus, ATCC), animal models (Mutant Mouse Resource & Research Centers, Mouse Genome Informatics), antibodies (Antibody Registry), genetic reagents (Addgene) and others (including SciCrunch, which aggregates information from several data sources). After selecting metadata attributes to collect for all tools as well as each specific type of tool, we constructed a relational data model for these attributes as well as other attributes that enable the inclusion of ‘observations’ for the tools, to be provided by database users.
A data model diagram for the NF Research Tools Database. Each box describes one of the database tables, with lines representing the relationships/connections between each table. Links between tables are the primary key (PK), which is a unique identifier for each row in the table, and the foreign key (FK), which is a PK found in another table. A machine-readable version of this data model is available on GitHub (see the ‘Database development’ section).
Database development
We constructed a JSON-LD schema from the data model to make it machine-readable (version 2.0.0 at the time of writing, available from https://github.com/nf-osi/nf-research-tools-schema). This schema was processed with the ‘schematic’ Python library (11, 12) to generate a MySQL database as well as data intake spreadsheets. Data were manually curated by searching for NF1-relevant tools on Scicrunch.org, Cellosaurus, Antibody Registry, Addgene, Mouse Genome Informatics and the Jackson Laboratories websites; this information was used to populate the data intake spreadsheets, which were then added to the MySQL database using schematic. The MySQL database is currently hosted in a private cloud computing environment, but all database tables and a variety of ‘joined’ tables are available for anyone to access on the Synapse collaborative science platform (www.doi.org/10.7303/syn26338068). The database tables can be retrieved and queried using SQL-like syntax in the Synapse web, Python, or R clients; please see the Synapse documentation (https://help.synapse.org) for more guidance on using these clients.
Web application
The web application was designed using the aforementioned user stories. We conducted internal design tests and solicited feedback from others at our organizations to improve the design. We then built the application using the React JavaScript library and the Synapse React Client (13).
Results
Search and explore tools
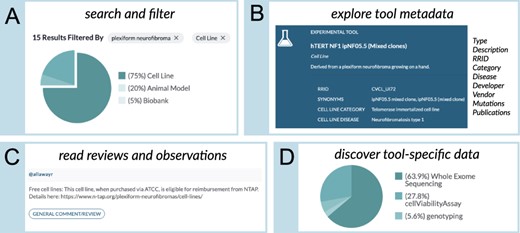
A central function of this database is to provide information about NF1-relevant research tools. In the web application, we included two avenues for exploring the tools listed in the database. From the home page, users can type in a search term of interest (e.g. ‘plexiform neurofibroma’; Figure 2A) to identify NF1-relevant animal models and cell lines. From there, users can use interactive, clickable charts to further filter and refine their results (e.g. limit results to cell lines; Figure 2A). The results of a specific, filtered search can then be shared with other researchers by sharing the URL or by downloading a table of the results.
The ‘Browse Tools’ user interface of the NF Data Portal allows researchers to interactively explore the NF Research Tools Database. (A) Clickable chart-based filters allow database users to filter tools using standardized metadata. Users can also search the tools by typing in free-text search terms. (B) The ‘Tool Detail’ pages allow users to perform a deep dive into information about specific research tools, for example, metadata describing the tool, the tool developer and other information described in panels (C, D). For example, a ‘canonical’ publication, when available, is listed for each tool. The Tool Detail pages also include publications that describe the use of the tool. When relevant information is available, the mutation/engineering status of selected genes—for example, genes that have been modified with the Cre-lox system or genes that have spontaneous mutations—is also provided. When tools are commercially available, the database provides a link to an external vendor, such as ATCC. (C) The Tool Details page also presents ‘Observations’, which are user-submitted details that describe pathology, usage notes, issues, reviews or other information relevant to a particular experimental tool. (D) When available in the NF Data Portal, data generated using a particular tool (e.g. publicly shared drug screening data generated using a commercially available cell line) are listed on the ‘Data’ tab of the Tool Detail page.
View tool details
The tools listed in the database have an associated ‘tool detail page’ (Figure 2B) that shows additional in-depth metadata, community-contributed observations and tool-related data (e.g. publications or data files generated using the research tool) available in the NF Data Portal. At the top, the detail page includes basic information such as the description, research resource identifier, research tool type-specific attributes (e.g. the category of cell line) and other metadata. These pages also include information about the developer of the research tool and, if available, information on publication(s) related to the development of the tool. When relevant to the tool type (i.e. predominantly animal models and cell lines), mutation information for selected genes (e.g. NF1) is presented and can be downloaded. When available, these pages also include a link to the vendor(s) of the research tool, as well as information about any usage restrictions, and whether a material transfer agreement is required.
This page also includes an ‘Observations’ tab (Figure 2C) that can display community-contributed observations such as the phenotypic timeline of a tool (e.g. developmental stages of a genetically engineered mouse model) as well as categorical observations such as pathologic observations, non-pathologic behavioral observations, neurochemical observations, comments from the depositing investigators, usage instructions, potential issues with the tool, reviews of the tool, general comments about the tool and other observations.
Finally, the tool detail pages contain a ‘Data’ tab (Figure 2D). This tab highlights data in the NF Data Portal that were generated using that tool, enabling, for example, users to find sequencing data or drug screening data for a cell line listed in the database.
Contribution to the database and other user support
We developed documentation for the database, which is available at help.nf.synapse.org. The ‘Neurofibromatosis Research Tools Database’ section of the documentation includes specific instructions for contributing new tools, contributing observations about a tool, as well as updating information about an existing tool. A core goal of this resource is to enable the community to contribute new tools and observations. We welcome any contributions for NF1, NF2 or schwannomatosis-relevant tools. For general help or issues that are not addressed by the documentation, our support team can be contacted by clicking on the ‘Need more help?’ button on the help.nf.synapse.org documentation page.
Conclusions
The NF Research Tools Database (tools.nf.synapse.org) was developed to mitigate challenges related to finding and accessing NF1-relevant research tools in a prepublication setting and sharing observations about how a tool behaves in real-world research settings. The NF research community is a small and highly collaborative community of researchers whose work often depends on the insights of researchers using the same tools; by providing a forum to share observations, we expect that NF researchers will be able to select, acquire and use research tools more easily. Furthermore, NF funding organizations like the Gilbert Family Foundation are attempting to expand the research community to non-NF1 researchers, and we anticipated that this database will lower the barrier of entry for these researchers to translate research advancements from other diseases to NF patients.
Currently, this prototype database includes a variety of NF1-relevant research tools. We are working on curating additional NF1-relevant tools, such as mouse models listed in Mutant Mouse Research and Resource Centers (MMRRC) (7) and the European Conditional Mouse Mutagenesis Program (14), and more prepublication models provided by the research community, as well as NF2- and schwannomatosis-related tools to increase the comprehensiveness of the database. In addition, we plan to add a guided form-based submission system so that users can more easily contribute standardized data to the database, tune the search feature to improve results and develop a process to share collected data, when possible, from this database with the respective RRID-minting authorities (e.g. Cellosaurus, AntibodyRegistry, Mouse Genome Informatics (MGI) and others) to expand the availability of this information in centralized tool-specific databases. Similarly, to increase the interoperability and utility of this database, we plan to expand our data model and add direct links to databases and external resources like ClinVar, MGI, Protein Data Bank and protocols.io. Finally, we plan to improve the user interface of the application by allowing users to search and filter tool observations and adding a ‘ratings’ system.
We also expect that this database and application could be used as an example to guide the development of other similar disease- or domain-specific research tool databases; both the data model and software are free, open-source and available for reuse by any interested communities. By collating, curating and surfacing this information in an open-access exploration tool, we anticipate that the NF Research Tools Database will be a valuable resource for the NF community.
Supplementary data
Supplementary data are available at Database Online.
Acknowledgements
The authors would like to thank Anh Nguyet Vu for data curation assistance, and Ryan Luce and Kaitlin Throgmorton for helpful conversations about this project.
Funding
Gilbert Family Foundation (821001); subset of the co-authors are employed by the Gilbert Family Foundation.
Conflict of interest
None declared.
Author contributions
Y.K., C.M., J.R.G. and R.J.A. were responsible for the conceptualization. https://credit.niso.org/implementing-credit/.
B.Z., M.D., J.R.G. and R.J.A. were responsible for the data curation. The formal analysis and investigation are not applicable in the article.
R.J.A. is involved in the funding acquisition.
J.A.E., M.D. and B.Z. were responsible for the methodology.
A.C., K.J.B., C.M., Y.K., R.J.A. and J.A.B. were responsible for the project administration.
J.R.G. and R.J.A. were responsible for the resources.
B.H., J.H., K.J.B., M.D., M.N., M.M. and S.S. were responsible for the software.
A.C., J.A.E., K.J.B., L.B., B.H., R.J.A. and J.A.B. were responsible for the supervision.
A.C., M.D., B.Z., C.M., K.V., Y.K., J.R.G. and R.J.A. were responsible for the validation.
J.H., S.S., L.B. and R.J.A. were responsible for the visualization.
A.C., C.M., J.A.E., J.B., K.V., J.R.G. and R.J.A. were responsible for the writing—original draft.
A.C., M.D., B.Z., J.H., C.M., S.S., J.A.E., M.N., J.B., K.V., M.M., K.J.B., B.H., L.B., Y.K., J.R.G., R.J.A. and J.A.B. were responsible for the writing—review & editing.
Note: J.B. is Jineta Banerjee; J.A.B. is Julie Bletz. All authors made a significant intellectual contribution to the project.
References
Author notes
contributed equally to this work.