-
PDF
- Split View
-
Views
-
Cite
Cite
Rezarta Islamaj, Robert Leaman, David Cissel, Cathleen Coss, Joseph Denicola, Carol Fisher, Rob Guzman, Preeti Gokal Kochar, Nicholas Miliaras, Zoe Punske, Keiko Sekiya, Dorothy Trinh, Deborah Whitman, Susan Schmidt, Zhiyong Lu, NLM-Chem-BC7: manually annotated full-text resources for chemical entity annotation and indexing in biomedical articles, Database, Volume 2022, 2022, baac102, https://doi.org/10.1093/database/baac102
Close - Share Icon Share
Abstract
The automatic recognition of chemical names and their corresponding database identifiers in biomedical text is an important first step for many downstream text-mining applications. The task is even more challenging when considering the identification of these entities in the article’s full text and, furthermore, the identification of candidate substances for that article’s metadata [Medical Subject Heading (MeSH) article indexing]. The National Library of Medicine (NLM)-Chem track at BioCreative VII aimed to foster the development of algorithms that can predict with high quality the chemical entities in the biomedical literature and further identify the chemical substances that are candidates for article indexing. As a result of this challenge, the NLM-Chem track produced two comprehensive, manually curated corpora annotated with chemical entities and indexed with chemical substances: the chemical identification corpus and the chemical indexing corpus. The NLM-Chem BioCreative VII (NLM-Chem-BC7) Chemical Identification corpus consists of 204 full-text PubMed Central (PMC) articles, fully annotated for chemical entities by 12 NLM indexers for both span (i.e. named entity recognition) and normalization (i.e. entity linking) using MeSH. This resource was used for the training and testing of the Chemical Identification task to evaluate the accuracy of algorithms in predicting chemicals mentioned in recently published full-text articles. The NLM-Chem-BC7 Chemical Indexing corpus consists of 1333 recently published PMC articles, equipped with chemical substance indexing by manual experts at the NLM. This resource was used for the evaluation of the Chemical Indexing task, which evaluated the accuracy of algorithms in predicting the chemicals that should be indexed, i.e. appear in the listing of MeSH terms for the document. This set was further enriched after the challenge in two ways: (i) 11 NLM indexers manually verified each of the candidate terms appearing in the prediction results of the challenge participants, but not in the MeSH indexing, and the chemical indexing terms appearing in the MeSH indexing list, but not in the prediction results, and (ii) the challenge organizers algorithmically merged the chemical entity annotations in the full text for all predicted chemical entities and used a statistical approach to keep those with the highest degree of confidence. As a result, the NLM-Chem-BC7 Chemical Indexing corpus is a gold-standard corpus for chemical indexing of journal articles and a silver-standard corpus for chemical entity identification in full-text journal articles. Together, these resources are currently the most comprehensive resources for chemical entity recognition, and we demonstrate improvements in the chemical entity recognition algorithms. We detail the characteristics of these novel resources and make them available for the community.
Database URL: https://ftp.ncbi.nlm.nih.gov/pub/lu/NLM-Chem-BC7-corpus/
Introduction
Chemical entities appear throughout the biomedical research literature, in studies from chemistry to various other disciplines such as medicine, biology and pharmacology. As such, chemical names are one of the most searched entity types in PubMed (1). Therefore, correctly identifying chemical names has a significant impact on chemical information retrieval: helping scientists retrieve the relevant literature, directly impacting research that relies on a correct understanding of the structure of chemicals, their usage and interactions with other molecular entities. For example, the correct identification of chemicals and their properties directly impacts drug development research (2).
However, chemicals in the biomedical literature often do not appear to conform to the chemical naming rules defined by standardization bodies. Chemicals appear in numerous lexical variations, synonymous names and abbreviated forms, which are often ambiguous (3). Moreover, these variations and difficulties are often compounded in the articles’ full text, compared with the title and abstract, causing a substantial performance reduction in automated chemical named entity recognition (NER) systems trained using only titles and abstracts (4). However, the full text frequently contains more detailed chemical information, such as the properties of chemical compounds, their biological effects and their interactions with diseases, genes and other chemicals (5–7).
Developing a chemical entity recognition system that accurately addresses these challenges requires a manually annotated corpus of chemical entities, with sufficient examples in full-text articles for system training and an accurate evaluation of its performance.
The NLM-Chem track at BioCreative VII (8) consisted of two tasks (9, 10):
Chemical Identification in full text: predicting all chemicals mentioned in recently published full-text articles, both span (i.e. NER) and normalization (i.e. entity linking), using Medical Subject Heading (MeSH; https://www.nlm.nih.gov/mesh/meshhome.html).
Chemical Indexing prediction task: predicting which chemicals mentioned in recently published full-text articles should be indexed, i.e. appear in the listing of MeSH terms for the document (11).
To support the challenge and address the need of creating high-quality chemical corpora, we introduced two rich and comprehensive chemical entity resources that contain manual annotations for chemical entities mentioned in articles’ text and manual indexing for the chemical substances that can represent an article’s topic and content. These resources are as follows:
The NLM-Chem-BC7 Chemical Identification Task corpus
The National Library of Medicine (NLM)-Chem BioCreative VII (NLM-Chem-BC7) Chemical Identification task corpus consists of 204 full-text PubMed Central (PMC) articles manually annotated for chemical entities by 12 NLM expert annotators. The first 150 articles, provided as the training set, were previously published as the NLM-Chem corpus (4), and the additional 54 full-text articles were specifically annotated for the BioCreative VII challenge and to serve as the Chemical Identification task testing set. Each article was doubly annotated in a three-round annotation process, where annotator discrepancies were discussed after each round until they reached full consensus. Finally, the articles were enriched with the manually indexed chemical substances.
The NLM-Chem-BC7 Chemical Indexing Task corpus
This collection consists of 1333 recently published (published in Spring 2021) full-text articles in the PMC Open Access collection, manually indexed with chemical substances. This set of articles was used as the testing set for the Chemical Indexing task, and the challenge participants were evaluated on the set of chemical indexing terms that were produced for these articles as part of the NLM’s regular operations.
Post challenge, the NLM-Chem track organizers worked in collaboration with 11 NLM indexers to validate the team submission results for article chemical indexing. As part of this work, chemical indexing terms that were given low prediction scores and indexing terms with high prediction scores that did not appear in the regular list of indexing terms were doubly reviewed in a blind experiment. During this validation experiment, the NLM indexers, unaware of how the terms were obtained, judged each term based on whether that term should be a topic description term for the corresponding article. We took this rare opportunity to show that the real value of automated methods is higher than that can be measured with current corpora. Article indexing at the NLM is performed by professional indexers who have years of experience categorizing and indexing the medical literature. Albeit even such highly experienced indexers sometimes do not agree 100%, and it is possible that different indexers could occasionally choose slightly different MeSH terms to reflect the topic terms of an article. This experiment allowed us to have the same article reviewed by two indexers in addition to the original indexer who provided the official indexing. As a result, we introduce a novel gold-standard resource, and the first of its kind, for article chemical indexing, the NLM-Chem-BC7 Chemical Indexing task corpus.
During the challenge, the participants in the Chemical Identification task returned chemical identification predictions for 1387 articles without knowing which articles would be used for the Identification task evaluation. After the challenge, we combined the submitted predictions (on the 1333 articles not used for evaluation) into an ensemble and created a silver-standard corpus for chemical identification. Our approach scored each span according to the proportion of the 53 valid submissions (plus the prediction from the original benchmark). We then applied a threshold and combined any remaining spans that overlapped. We finally normalized each span by determining the identifier most frequently associated with the associated <document ID, mention text> pair.
In addition, the NLM-Chem track in BioCreative VII also presented an extended chemical entity annotated collection from previous BioCreative challenges, utilizing the chemical entity corpora from previous BioCreative challenges [CHEMDNER (3) and BC5CDR (12)]. The articles in these corpora were enriched with the NLM MeSH chemical substance indexing by the challenge organizers and converted into the same format (BioC XML and JSON) as the NLM-Chem datasets. The goal is to provide a continuity of chemical entity identification research and promote data reuse.
The NLM-Chem-BC7 corpora contain, respectively, 204 and 1333 full-text PMC Open Access articles and differ from previous corpora because the articles were selected to be rich in chemical mentions, rich in other biomedical entities and representative for current research on chemicals and drugs. These articles were published in many different journals to represent a large space of language variation. Most importantly, both resources can be combined with the previously published chemical entity annotation resources to facilitate research. We have validated the utility of this corpus, and we believe the availability of this corpus will foster newer developments for more accurate chemical entity prediction algorithms. These characteristics make them invaluable for the advancement and improvement of text-mining tools for accurate chemical entity identification and chemical entity article indexing (topic prediction).
The data can be found at https://ftp.ncbi.nlm.nih.gov/pub/lu/NLM-Chem-BC7-corpus/.
Methods
Document selection procedure
The chemical corpus of the NLM-Chem BioCreative VII track had these targets:
Be representative of biomedical literature publications that contain chemical mentions.
Target articles for which human annotation was most valuable.
Be instrumental in training chemical NER algorithms to produce high-quality results in full-text publications, as well as article abstracts.
To select candidate articles for human annotation for the NLM-Chem-BC7 corpus, we evaluated each article as follows:
To be rich in chemical entities that current NER tools have trouble identifying.
To have no restrictions on sharing and distribution.
To be useful for other downstream biomedical entity text-mining-related tasks.
To select the articles most suitable for algorithm testing, in addition to the constraints above, we focused on recently published articles. Chemical NER and indexing algorithms are most valuable for the incoming flux of published literature. As we experienced with the coronavirus disease pandemic, correctly identifying chemicals and drugs discussed in the articles, as well as grouping those articles by the relevant substances, is most crucial, especially in the race to find an effective cure and a timely vaccine.
The 54 full-text articles that constituted the Chemical Identification task testing set were selected to be as similar as possible to the NLM-Chem corpus of 150 full-text articles (4), to be complementary, balancing and a suitable test set, that can also serve as a stand-alone corpus. The selection criteria included maximization of journal coverage to assure variety, similar distribution of chemical mentions and identifiers per article, similar distribution of other biomedical entities per article and similar language models.
We repurposed the CHEMDNER and the BC5CDR corpora for the NLM-Chem track challenge. The CHEMDNER documents are title/abstract annotations for chemical NER and do not include the chemical normalization. However, as this could still be useful for deep learning strategies, we converted all the articles and their annotations in the same format as NLM-Chem corpus documents. The BC5CDR corpus, on the other hand, contains title/abstract chemical annotations and their MeSH identifiers; we therefore converted these documents in the same format.
We filtered the manual MeSH indexing terms assigned to each article in the MEDLINE collection at the NLM to extract the chemical substances to support the Chemical Indexing task. These indexing terms represent chemical substances that are important topics in their respective articles and, therefore, are valuable for chemical information retrieval. We extracted the indexed chemical substances and enriched the dataset for every article in the NLM-Chem, CHEMDNER and BC5CDR corpora.
The Chemical Indexing testing set consisted of 1333 recently published articles and was selected using the same criteria for the Chemical Identification testing set. These articles were manually indexed with MeSH indexing terms, as per the routine NLM operations, after the completion of the NLM-Chem track challenge, during September 2021, and those indexed labels were used for the BioCreative VII NLM-Chem Indexing task evaluation. After the challenge, these articles were further used to create the gold- and silver-standard corpora.
Chemical entity annotation guidelines in the full text
The complete NLM-Chem corpus annotation guidelines are publicly available with the corpus (4). We followed the same guidelines, with the same group of NLM professional indexers as annotators, and here we give a quick summary.
Our guidelines specify which text elements should be tagged, those that should not be tagged and how to assign the tagged mentions to their corresponding MeSH identifiers. The primary considerations of the annotation guidelines are (i) what should be labeled as a chemical, (ii) how to place the mention boundaries for those labels and (iii) how to associate those mentions with an entity within one of the chemical trees of MeSH.
Creating high-quality guidelines that fit the annotation task required a multistep iterative process, starting from an initial draft that was revised several times until clear and refined guidelines were obtained. We found that defining the text-bound annotations of chemical mentions found in full-text articles was not trivial. It required a deep knowledge of chemistry, supported by the consultation of external knowledge sources (the MeSH vocabulary, PubChem, etc.). The guidelines were prepared by 12 professional MeSH indexers with degrees in chemistry, biochemistry, biological sciences and molecular biology and an average of 20 years of experience in indexing PubMed literature with MeSH indexing terms.
First, it was decided that very general chemical concepts [such as atom(s) and moiety (moieties)] and terms that cannot be associated directly with a chemical structure such as molecule(s), drug(s) and polymer(s) should be excluded from the annotation. In addition, macromolecular biochemicals, namely, proteins (including enzymes), lipids and nucleic acids (DNA and RNA), were excluded from annotation. In addition, embedded chemical concepts in other biomedical entities such as ‘sodium channel gene’, where the chemical concept ‘sodium’ is embedded in a phrase indicating a different type of biochemical entity ‘gene’, were tagged as OTHER. Each rule defined in the guidelines was also represented by one or more illustrative examples to simplify comprehension and application. (Please see Supplementary Materials for a copy of the annotation guidelines for chemical entities in full text).
Annotation procedure for the NLM-Chem-BC7 Chemical Identification task corpus
The NLM-Chem-BC7 full-text articles are doubly annotated by 12 NLM experts in three annotation rounds using the TeamTat annotation tool (13). All articles were pre-annotated using the NLM-Chem improved chemical recognition tool, with performances not far from the human inter-annotator agreement values, measuring 76% and 77% in f-measure for chemical name entity recognition and chemical normalization, respectively (4). Articles were randomly assigned to pairs of annotators in such a way that the annotation burden was equally distributed. The first round of manual annotations consisted of each annotator working on and completing the annotations of the assigned articles independently. At this stage, the annotators did not know the identities of their partners. After completion, these annotations were reviewed by the technical team to identify differences and discrepancies. Inter-annotator agreement was measured to be 68%. All pairwise annotations were merged into one document, and the agreements and disagreements were marked and made available in the annotation tool for annotation Round 2. The second round of annotations consisted of each annotator working independently in their own annotation space, again without knowing the identities of their partner annotators. They reviewed their own decisions and considered their partners’ decisions editing the documents until they were satisfied. After completion, the annotations were again reviewed, inter-annotator agreement was computed to be 84% and the remaining differences and discrepancies were analyzed. All annotations were again merged into one document, the agreements and remaining disagreements were marked and the documents were made available to the respective annotators’ accounts. In the third and final round of annotations, the annotation partners for each document were revealed, and every pair of annotators collaboratively reviewed and discussed any remaining differences and finalized the shared document annotation reaching 100% complete consensus.
Document format
While annotations can be represented in various formats, we used the BioC (XML and JSON) format due to several considerations: (i) the format (14) supports full-text articles and annotations representing both mention span (location) and entity identifier; (ii) articles in the PMC text-mining subset (15) are already available in BioC; (iii) our annotation tool of choice TeamTat and the NLM-Chem NER tool already support the format and (iv) the format is simple and easy to modify, allowing additional analysis tools to be applied rapidly as needed.
The NLM-Chem-BC7 gold and silver corpus development
The challenge participants of the NLM-Chem track submitted prediction results (8) for all the 1387 full-text articles for both chemical entity prediction and chemical indexing tasks, as they were not aware which articles constituted the Chemical Identification task testing dataset. After the BioCreative VII challenge, the NLM-Chem track organizers reviewed all challenge submission results and developed the Chemical Indexing gold-standard corpus and the Chemical Identification silver-standard corpus, as follows:
The NLM-Chem-BC7 Chemical Indexing gold-standard corpus
We received 18 submissions for the Chemical Indexing task, and all of them were considered. We selected the terms for manual validation from the team submissions using a probabilistic classification approach by estimating the probability that a predicted MeSH indexing term is a true term. We built a naïve Bayes classifier using each submission as a binary feature, determining the classifier weights directly (without training) from the actual accuracy of each submission compared with the original MeSH indexing.
To estimate the probability of a given MeSH indexing term, we first gather the labels for that term across all submissions as |${{\rm{l}}_{\rm{i}}} \in \left\{ { - 1,{\ }1} \right\}$|, where 1 indicates that the submission predicts the specified term and −1 indicates that the submission does not predict the specified term. Then we estimate the probability of the specified MeSH term as follows:
We aggregated all terms predicted by any submission and all terms from the original MeSH indexing and associated with each term a probability of being a true MeSH indexing term based on the predicted output of the classifier. We then determined which terms should be manually validated by thresholding. Specifically, MeSH terms were manually validated if they were (i) not part of the original MeSH indexing but were given a score greater than 0.1—meaning possibly useful—or (ii) MeSH terms part that were included in the original MeSH indexing but were given a score less than 0.5—meaning less likely to be predicted by the automatic classifiers. Note that we also considered a confidence score approach where each term received a score reflecting the proportion of the submissions that predicted the term. We found the two approaches to behave similarly when the majority of submissions included (or excluded) a term; however, using the actual accuracy of the submission allowed for improving predictions in cases of disagreement.
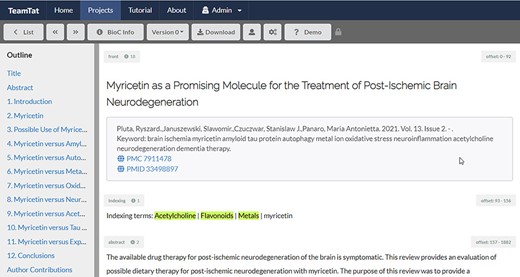
The predicted list of the chemical substances was compared with the official NLM list of indexed chemical substances, extracted from the metadata of the articles downloaded on 30 September 2021. All discrepancies, indexed chemical substances not appearing in the predicted substances list and all predicted substances (scoring above a threshold) not appearing in the indexed substances list were compiled for each article. These articles were organized as part of a new annotation project on the TeamTat annotation tool. All articles were uploaded to TeamTat divided into nine batches containing 125–127 articles each and were distributed to 11 NLM indexers. Each indexer reviewed 209–210 articles in total for chemical indexing terms during January and February 2022, so that each article was annotated by two indexers. All articles were randomly distributed, so that the number of articles reviewed by every pair of indexers was equally distributed. The indexers worked in two rounds: during Round 1 they worked independently and selected the indexing terms and during Round 2 they worked in pairs and resolved any remaining discrepancies. The inter-annotator agreement after Round 1 was 73%. After Round 1, annotators discussed the remaining disagreements and came to a 100% consensus. It is important to note that a disagreement generally consisted of a discussion whether specific indexing rules applied such as this one: if three or more specific concept terms (chemicals) are mentioned in the article, that article is indexed with a parent concept term that covers the specific terms. Figure 1 shows a screenshot of a random article and illustrates how this task was performed.
Illustration of indexing review in TeamTat. Every article was reviewed by two indexers. The image shows a screenshot of the article and a section under the article title named ‘Indexing terms’. All the chemical terms to be reviewed for each article are listed in this section. The NLM indexers highlighted the chemical indexing terms that best represented the article topics.
The NLM-Chem-BC7 Chemical Identification silver-standard corpus
All 53 valid submissions for the Chemical Identification task, plus the original benchmark system, were used to create the NLM-Chem silver-standard corpus. Our approach uses two phases corresponding to NER and normalization. For NER, we gathered all annotated text mentions (spans) from the submissions and scored each mention according to the proportion of the prediction sets that contain the mention. We used the 54 articles from the testing set to identify the threshold that maximizes the F-score and applied that threshold to the 1333 articles in the silver-standard set. Any remaining mentions that overlapped were then combined. The mentions were then normalized by determining the identifier most frequently associated with the associated <document ID, mention text> pair across all submissions.
Results
Corpus characteristics for chemical identification
The NLM-Chem track chemical resources are rich in manual chemical annotations. The NLM-Chem-BC7 Chemical Identification corpus is currently the largest manually annotated corpus of full-text journal articles targeted for developing chemical NER text-mining tools and is compatible with previously annotated corpora. The NLM-Chem-BC7 Chemical Identification corpus training dataset (4) consists of 150 full-text articles containing 38 339 manual chemical mention annotations, corresponding to 4862 unique chemical name strings, normalized to 1810 MeSH identifiers. The NLM-Chem-BC7 Chemical Identification corpus testing dataset consists of 54 recently published full-text articles, containing 22 942 manual chemical mention annotations, corresponding to 3419 unique chemical strings and 1352 unique MeSH IDs. The BC5CDR set contains 15 951 chemical mention annotations, corresponding to 2639 unique chemical name strings, normalized to 1269 MeSH identifiers. The CHEMDNER set contains 84 331 chemical mention annotations, corresponding to 19 803 unique chemical name strings. The NLM-Chem-BC7 Chemical Identification silver-standard corpus consists of 1333 recently published full-text articles, automatically annotated for all occurrences of chemical entities in the full text. This dataset contains 392 838 automatic chemical mention annotations, corresponding to 33 209 unique chemical strings and 12 301 unique MeSH IDs.
The NLM-Chem-BC7 Chemical Indexing gold-standard corpus of 1333 recently published full-text articles contains 1798 unique indexed chemical terms (MeSH IDs). The statistics of annotations per article per dataset are detailed in Table 1.
Data characteristics of the NLM-Chem track datasets
| . | NLM-Chem-BC7 Identification . | NLM-Chem-BC7 Indexinga . | BC5CDR . | CHEMDNER . |
|---|---|---|---|---|
| Number of articles | 204 (full text) | 1333 (full text) | 1500 (abstract) | 10 000 (abstract) |
| Number of chemical annotations per article (unique) | ||||
| Minimum | 2 (1) | 2 (1) | 1 (1) | 0 (0) |
| Maximum | 1318 (214) | 1924 (412) | 55 (22) | 67 (40) |
| Average | 300.4 (66.6) | 294.7 (66.8) | 10.6 (4.1) | 8.4 (4.6) |
| Median | 279 (60) | 262 (58) | 9 (3) | 7 (4) |
| Number of unique MeSH identifiers per article | ||||
| Minimum | 1 | 1 | 1 | NA |
| Maximum | 127 | 244 | 16 | NA |
| Average | 40.5 | 45.6 | 2.9 | NA |
| Median | 39.0 | 41.0 | 2 | NA |
| Number of unique indexed substances per article | ||||
| Minimum | 0 | 0 | 0 | 0 |
| Maximum | 14 | 14 | 11 | 19 |
| Average | 1.8 | 3.2 | 2.3 | 2.2 |
| Median | 1 | 3 | 2 | 2 |
| . | NLM-Chem-BC7 Identification . | NLM-Chem-BC7 Indexinga . | BC5CDR . | CHEMDNER . |
|---|---|---|---|---|
| Number of articles | 204 (full text) | 1333 (full text) | 1500 (abstract) | 10 000 (abstract) |
| Number of chemical annotations per article (unique) | ||||
| Minimum | 2 (1) | 2 (1) | 1 (1) | 0 (0) |
| Maximum | 1318 (214) | 1924 (412) | 55 (22) | 67 (40) |
| Average | 300.4 (66.6) | 294.7 (66.8) | 10.6 (4.1) | 8.4 (4.6) |
| Median | 279 (60) | 262 (58) | 9 (3) | 7 (4) |
| Number of unique MeSH identifiers per article | ||||
| Minimum | 1 | 1 | 1 | NA |
| Maximum | 127 | 244 | 16 | NA |
| Average | 40.5 | 45.6 | 2.9 | NA |
| Median | 39.0 | 41.0 | 2 | NA |
| Number of unique indexed substances per article | ||||
| Minimum | 0 | 0 | 0 | 0 |
| Maximum | 14 | 14 | 11 | 19 |
| Average | 1.8 | 3.2 | 2.3 | 2.2 |
| Median | 1 | 3 | 2 | 2 |
Chemical indexing statistics refer to the gold corpus, and chemical annotation statistics refer to the silver corpus.
Data characteristics of the NLM-Chem track datasets
| . | NLM-Chem-BC7 Identification . | NLM-Chem-BC7 Indexinga . | BC5CDR . | CHEMDNER . |
|---|---|---|---|---|
| Number of articles | 204 (full text) | 1333 (full text) | 1500 (abstract) | 10 000 (abstract) |
| Number of chemical annotations per article (unique) | ||||
| Minimum | 2 (1) | 2 (1) | 1 (1) | 0 (0) |
| Maximum | 1318 (214) | 1924 (412) | 55 (22) | 67 (40) |
| Average | 300.4 (66.6) | 294.7 (66.8) | 10.6 (4.1) | 8.4 (4.6) |
| Median | 279 (60) | 262 (58) | 9 (3) | 7 (4) |
| Number of unique MeSH identifiers per article | ||||
| Minimum | 1 | 1 | 1 | NA |
| Maximum | 127 | 244 | 16 | NA |
| Average | 40.5 | 45.6 | 2.9 | NA |
| Median | 39.0 | 41.0 | 2 | NA |
| Number of unique indexed substances per article | ||||
| Minimum | 0 | 0 | 0 | 0 |
| Maximum | 14 | 14 | 11 | 19 |
| Average | 1.8 | 3.2 | 2.3 | 2.2 |
| Median | 1 | 3 | 2 | 2 |
| . | NLM-Chem-BC7 Identification . | NLM-Chem-BC7 Indexinga . | BC5CDR . | CHEMDNER . |
|---|---|---|---|---|
| Number of articles | 204 (full text) | 1333 (full text) | 1500 (abstract) | 10 000 (abstract) |
| Number of chemical annotations per article (unique) | ||||
| Minimum | 2 (1) | 2 (1) | 1 (1) | 0 (0) |
| Maximum | 1318 (214) | 1924 (412) | 55 (22) | 67 (40) |
| Average | 300.4 (66.6) | 294.7 (66.8) | 10.6 (4.1) | 8.4 (4.6) |
| Median | 279 (60) | 262 (58) | 9 (3) | 7 (4) |
| Number of unique MeSH identifiers per article | ||||
| Minimum | 1 | 1 | 1 | NA |
| Maximum | 127 | 244 | 16 | NA |
| Average | 40.5 | 45.6 | 2.9 | NA |
| Median | 39.0 | 41.0 | 2 | NA |
| Number of unique indexed substances per article | ||||
| Minimum | 0 | 0 | 0 | 0 |
| Maximum | 14 | 14 | 11 | 19 |
| Average | 1.8 | 3.2 | 2.3 | 2.2 |
| Median | 1 | 3 | 2 | 2 |
Chemical indexing statistics refer to the gold corpus, and chemical annotation statistics refer to the silver corpus.
Figure 2 illustrates that these resources are (i) compatible—to foster reuse, acknowledge and build on previous efforts of experts and (ii) complementary—to expand on previous knowledge and cover new areas of training data. Furthermore, Fig. 2 illustrates the impact of annotations in the full text. As seen, the full text contains much more chemical annotations and a larger variety both in the mention as well as the respective identifiers. The NLM-Chem-BC7 manually annotated data in full-text articles allows the new algorithms to learn from and explore a space of chemical mentions in the biomedical literature that had not been covered in previously annotated corpora, as illustrated with the overlap with the BC5CDR and CHEMDNER corpora. Finally, NLM-Chem-BC7 and the BC5CDR corpora contain chemical annotations normalized to MeSH identifiers, which, via Unified Medical Language System (UMLS, https://www.nlm.nih.gov/research/umls/index.html), can be mapped to different chemical terminologies as needed.
Illustration of common and different chemical annotations in the chemical annotated data [The overlap of chemical mention annotations is shown in (a), the overlap of MeSH ID annotations is shown in (b) and the overlap of the indexed chemical substances is shown in (c)]. Please note that this depiction does not preserve the relative size of corpora. The NLM-Chem dataset of full-text articles brings in additional previously unseen chemical mentions and chemical identifiers.
Corpus characteristics for chemical indexing
In addition to the resources for chemical identification, NLM-Chem track provided novel resources, previously not available for text mining, manually verified for chemical indexing of journal articles. The three resources described above for chemical identification (NLM-Chem-BC7 Chemical Identification, CHEMDNER and BC5CDR) have been enriched with the MeSH-indexed chemical substances, representing chemical topic terms, opening up new research avenues in chemical information retrieval. Table 1 and Fig. 2 show the indexing terms for the chemical substances identified in each of the corpora discussed above.
In order to provide a better, larger and more comprehensive resource for chemical indexing, the NLM-Chem track organizers worked on the NLM Chemical Indexing gold-standard corpus. This corpus consists of 1333 full-text articles, from 61 journals, published in Spring 2021, and contains 1798 unique chemical indexing terms. In addition, these articles have been automatically annotated for chemical identification using the Chemical Identification task submissions as an ensemble to create a silver-standard dataset. The silver-standard corpus contains 392 838 total annotations, corresponding to 33 209 unique chemical mentions. These mentions are mapped to 12 301 unique MeSH ID identifiers, of which 10 055 (85.6%) do not appear in the BC5CDR or NLM-Chem-BC7 Identification corpora.
The most important contribution is that after the BioCreative VII we designed a chemical indexing validation experiment to evaluate the utility of the automatically predicted indexing terms. Table 2 and Table 3 summarize the results of this validation experiment. As shown in Table 2, the indexers removed 18.63% of the previously indexed terms; however, they accepted and added to the chemical indexing terms list 73.73% of the automatically suggested chemical substance terms. Upon further analysis, we verified that the algorithmically predicted terms are mainly specific chemical substances, while the indexer-supplied terms are mainly MeSH descriptor terms. This difference is important and necessary to note because this implies that automatic chemical indexing could significantly help human indexing by extracting with high accuracy the substances described in each article. Given these data, when we evaluate the exact match between algorithm predictions and human indexing, the current prediction accuracy is in the low 40%; however, when we adjust for topic descriptors, which fall higher in the MeSH hierarchy, the accuracy rises to almost 80%. Based on these results, the automatic chemical substance prediction could be accurate enough to be implemented as a chemical indexing precursor step, followed by additional steps to suggest the corresponding descriptor terms. Then, as necessary, the data could be presented to a human indexer to decide the final list of topic terms for article indexing, thus saving valuable human expert time.
Results of the chemical indexing validation experiment, where indexers blindly reviewed a set of terms that were present in the indexing metadata (but not algorithmically predicted) and terms that were suggested by the prediction algorithms but not indexed
| . | Final annotation . | . | |
|---|---|---|---|
| . | Yes . | No . | Total . |
| Indexed not predicted | 1930 (81.37%) | 442 (18.63%) | 2372 |
| Algorithm suggested, not indexed | 798 (73.73%) | 284 (26.25%) | 1082 |
| Total | 2728 | 726 | |
| . | Final annotation . | . | |
|---|---|---|---|
| . | Yes . | No . | Total . |
| Indexed not predicted | 1930 (81.37%) | 442 (18.63%) | 2372 |
| Algorithm suggested, not indexed | 798 (73.73%) | 284 (26.25%) | 1082 |
| Total | 2728 | 726 | |
Results of the chemical indexing validation experiment, where indexers blindly reviewed a set of terms that were present in the indexing metadata (but not algorithmically predicted) and terms that were suggested by the prediction algorithms but not indexed
| . | Final annotation . | . | |
|---|---|---|---|
| . | Yes . | No . | Total . |
| Indexed not predicted | 1930 (81.37%) | 442 (18.63%) | 2372 |
| Algorithm suggested, not indexed | 798 (73.73%) | 284 (26.25%) | 1082 |
| Total | 2728 | 726 | |
| . | Final annotation . | . | |
|---|---|---|---|
| . | Yes . | No . | Total . |
| Indexed not predicted | 1930 (81.37%) | 442 (18.63%) | 2372 |
| Algorithm suggested, not indexed | 798 (73.73%) | 284 (26.25%) | 1082 |
| Total | 2728 | 726 | |
Results of the chemical indexing validation experiment
| . | Final data summary . |
|---|---|
| Previously indexed and predicted | 1606 |
| Newly indexed and predicted | 798 |
| Indexed and not predicted | 1930 |
| Not indexed and predicted | 284 |
| Total predicted | 2687 |
| Total indexed | 4334 |
| . | Final data summary . |
|---|---|
| Previously indexed and predicted | 1606 |
| Newly indexed and predicted | 798 |
| Indexed and not predicted | 1930 |
| Not indexed and predicted | 284 |
| Total predicted | 2687 |
| Total indexed | 4334 |
Results of the chemical indexing validation experiment
| . | Final data summary . |
|---|---|
| Previously indexed and predicted | 1606 |
| Newly indexed and predicted | 798 |
| Indexed and not predicted | 1930 |
| Not indexed and predicted | 284 |
| Total predicted | 2687 |
| Total indexed | 4334 |
| . | Final data summary . |
|---|---|
| Previously indexed and predicted | 1606 |
| Newly indexed and predicted | 798 |
| Indexed and not predicted | 1930 |
| Not indexed and predicted | 284 |
| Total predicted | 2687 |
| Total indexed | 4334 |
Corpus technical validation
Table 4 shows the results of our benchmark method on the Chemical Identification task. This benchmark is based on our previously published method and is currently our best-performing chemical NER tool. This tool is used in the daily processing of the PubMed and PMC articles as they are queries in our PubTator Central portal (16). This implementation was trained only on the NLM-Chem full-text articles as the training dataset and tested on the NLM-Chem Chemical Identification task (50 full-text articles) dataset. Table 4 also shows results when the information contained in the silver-standard chemical name annotations were included in the training data. As expected, we see a significant improvement in performance. Given the enrichment in chemicals that we observe when we consider the biomedical articles’ space covered with the addition of BC5CDR and CHEMDNER corpora, it is reasonable to expect a further improvement in the chemical entity recognition in biomedical articles.
Benchmark results of the Chemical Identification task
| . | Chemical entity recognition . | |||||
|---|---|---|---|---|---|---|
| . | Strict . | Relaxed . | ||||
| Training dataset . | Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| NLM-Chem full-text articles | 0.8580 | 0.8383 | 0.8480 | 0.9221 | 0.8974 | 0.9096 |
| + NLM-Chem-BC7 silver corpus | 0.8767 | 0.8530 | 0.8647 | 0.9313 | 0.9071 | 0.9190 |
| Chemical entity normalization | ||||||
| NLM-Chem full-text articles | 0.8729 | 0.7641 | 0.8149 | 0.8630 | 0.7732 | 0.8122 |
| + NLM-Chem-BC7 silver corpus | 0.8919 | 0.7610 | 0.8213 | 0.8792 | 0.7705 | 0.8213 |
| . | Chemical entity recognition . | |||||
|---|---|---|---|---|---|---|
| . | Strict . | Relaxed . | ||||
| Training dataset . | Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| NLM-Chem full-text articles | 0.8580 | 0.8383 | 0.8480 | 0.9221 | 0.8974 | 0.9096 |
| + NLM-Chem-BC7 silver corpus | 0.8767 | 0.8530 | 0.8647 | 0.9313 | 0.9071 | 0.9190 |
| Chemical entity normalization | ||||||
| NLM-Chem full-text articles | 0.8729 | 0.7641 | 0.8149 | 0.8630 | 0.7732 | 0.8122 |
| + NLM-Chem-BC7 silver corpus | 0.8919 | 0.7610 | 0.8213 | 0.8792 | 0.7705 | 0.8213 |
Benchmark results of the Chemical Identification task
| . | Chemical entity recognition . | |||||
|---|---|---|---|---|---|---|
| . | Strict . | Relaxed . | ||||
| Training dataset . | Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| NLM-Chem full-text articles | 0.8580 | 0.8383 | 0.8480 | 0.9221 | 0.8974 | 0.9096 |
| + NLM-Chem-BC7 silver corpus | 0.8767 | 0.8530 | 0.8647 | 0.9313 | 0.9071 | 0.9190 |
| Chemical entity normalization | ||||||
| NLM-Chem full-text articles | 0.8729 | 0.7641 | 0.8149 | 0.8630 | 0.7732 | 0.8122 |
| + NLM-Chem-BC7 silver corpus | 0.8919 | 0.7610 | 0.8213 | 0.8792 | 0.7705 | 0.8213 |
| . | Chemical entity recognition . | |||||
|---|---|---|---|---|---|---|
| . | Strict . | Relaxed . | ||||
| Training dataset . | Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| NLM-Chem full-text articles | 0.8580 | 0.8383 | 0.8480 | 0.9221 | 0.8974 | 0.9096 |
| + NLM-Chem-BC7 silver corpus | 0.8767 | 0.8530 | 0.8647 | 0.9313 | 0.9071 | 0.9190 |
| Chemical entity normalization | ||||||
| NLM-Chem full-text articles | 0.8729 | 0.7641 | 0.8149 | 0.8630 | 0.7732 | 0.8122 |
| + NLM-Chem-BC7 silver corpus | 0.8919 | 0.7610 | 0.8213 | 0.8792 | 0.7705 | 0.8213 |
Table 5 shows the results of our baseline method on the Chemical Indexing task. For this task, we added a component to our Chemical Identification benchmark to return the set of MeSH identifiers from annotations found in the title and abstract as the set of indexed chemicals. The indexing component thus represents a straightforward baseline approach with relatively low precision but higher recall.
Benchmark results Chemical Indexing task
| Chemical indexing terms prediction . | |||||
|---|---|---|---|---|---|
| Strict . | Relaxed . | ||||
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| 0.7351 | 0.5858 | 0.6520 | 0.7920 | 0.7450 | 0.7277 |
| Chemical indexing terms prediction . | |||||
|---|---|---|---|---|---|
| Strict . | Relaxed . | ||||
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| 0.7351 | 0.5858 | 0.6520 | 0.7920 | 0.7450 | 0.7277 |
Benchmark results Chemical Indexing task
| Chemical indexing terms prediction . | |||||
|---|---|---|---|---|---|
| Strict . | Relaxed . | ||||
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| 0.7351 | 0.5858 | 0.6520 | 0.7920 | 0.7450 | 0.7277 |
| Chemical indexing terms prediction . | |||||
|---|---|---|---|---|---|
| Strict . | Relaxed . | ||||
| Precision . | Recall . | F-score . | Precision . | Recall . | F-score . |
| 0.7351 | 0.5858 | 0.6520 | 0.7920 | 0.7450 | 0.7277 |
The strict evaluation for both chemical entity recognition and normalization tasks assumes an exact match between the predicted mention span or MeSH identifier and the annotated mention span or MeSH identifier. The relaxed evaluation for chemical entity recognition considers a predicted mention span to match an annotated mention span if they overlap. For chemical entity normalization, which is evaluated both in the Chemical Identification task and the Chemical Indexing task, the relaxed evaluation is the least common ancestor F-score (17). Please note that in the case of relaxed evaluation, the least common ancestor consideration introduces both more possible candidate entities and more available entities overall, and as a result, the F-score could be lower.
Conclusions
The NLM-Chem BioCreative track developed important high-quality chemical resources, of value to the community, which are as follows:
The NLM-Chem-BC7 Chemical Identification corpus, which consists of 204 full-text articles doubly annotated by 12 NLM indexers in three rounds of annotation, reaching full consensus and resolving any annotator disagreements. This corpus is currently the largest corpus of full-text articles annotated with chemical entities at a high degree of granularity and their NLM-indexed chemical substances. The NLM-Chem training dataset (150 articles) contains a total of 38 339 manual chemical mention annotations, corresponding to 4862 unique chemical name strings, normalized to 1810 MeSH identifiers. The NLM-Chem Chemical Identification testing dataset (50 articles) contains 3740 unique chemical strings and 1352 unique MeSH IDs. The articles were carefully selected from the PMC Open Access dataset and covered 71 journals.
The extended chemical entity annotated collection from previous BioCreative challenges (CHEMDNER and BC5CDR). These articles were enriched with the manually indexed chemical substances.
The NLM-Chem-BC7 Chemical Indexing corpus. This resource consists of 1333 recently published full-text articles in the PMC Open Access collection, manually indexed with chemical substances. This set of articles was used as the testing set for the Chemical Indexing task. This set was further refined to serve as a gold-standard corpus for chemical indexing and silver-standard corpus for chemical annotation in full-text biomedical journal articles.
For the chemical indexing purpose, 11 NLM indexers reviewed in a blind experiment all articles’ chemical indexing terms that were not predicted in any of the submitted results by the participating teams and the chemical indexing terms predicted by the majority of teams, but not included in the articles’ indexing metadata. This experiment revealed that most algorithm-predicted terms would be selected for indexing when reviewed by a human expert (73.73%) and that most current indexing terms would be again selected for indexing (81.36%). These results reflect the differences between entity identification and topic identification tasks—while chemical entity prediction algorithms are focused on identifying specific substances, additional steps utilizing the MeSH hierarchy are necessary to supply all the related topic terms For the chemical annotation purpose, all chemical entity annotations submitted from all teams participating in the challenge were merged via a linear programming approach and they were combined into a silver-standard corpus. This corpus was used for retraining the initial benchmark, and we saw marked improvements, especially in the chemical mention recognition step.
To provide a robust test of the corpus utility in chemical entity recognition and normalization that could translate to real-life applications, we tested the new corpus with our best-performing chemical NER and normalization tool, based on a deep learning architecture for the name entity recognition component and a multi-terminology candidate resolution architecture for the normalization component.
The NLM-Chem track chemical resources provide these contributions: (i) high-quality manual annotation of chemical entities in the full text, (ii) chemical entity normalization to MeSH identifiers, which via UMLS, can be easily mapped to other chemical terminologies, if needed, and (iii) chemical terms indexing of all articles, representing the chemical topic terms for these articles as indexed by the expert literature indexers at the NLM. The annotation guidelines are compatible with previously annotated corpora; therefore, the previous (abstract-only) corpora can be used as additional data. The enriched chemical resource of the NLM-Chem track challenge will be invaluable for advancing text-mining techniques for chemical extraction tasks in biomedical text.
Supplementary Material
Supplementary Material is available at Database online.
Data Availability
The corpus described in this study is available at: https://ftp.ncbi.nlm.nih.gov/pub/lu/NLM-Chem-BC7-corpus/.
Funding
National Institues of Health (NIH) Intramural Research Program, National Library of Medicine.
Conflict of interest
None declared.
References
Author notes
Co-first authors



![Illustration of common and different chemical annotations in the chemical annotated data [The overlap of chemical mention annotations is shown in (a), the overlap of MeSH ID annotations is shown in (b) and the overlap of the indexed chemical substances is shown in (c)]. Please note that this depiction does not preserve the relative size of corpora. The NLM-Chem dataset of full-text articles brings in additional previously unseen chemical mentions and chemical identifiers.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/database/2022/10.1093_database_baac102/2/m_baac102f2.jpeg?Expires=1716727509&Signature=jRn9M-o2rGGP3ZcwQqpGWIXsTQtSzRv0Vax-vb41p8VZW~qWDBiE5Jr85NJEmmRImTviTf3nVyRduEWnngG-3QJfXDqwu4NubgV4VmNE0KMgcqnzsENjBBpFbkB3VYjIF2U20wEyz~~nWZyCeeD8zoOx1mCq1eKvJjVecftY2mazIww3SDeE63MLBWsj~c22vCi5q3qr2cIE61q1RcfbLS9dMJGNlpeCgHyAF9qmYfcxCf7OyMWPh9OKwvRj33wV4BgmBFFUGULKAKjS8iazmtxKq1jgIVWGjag4wTiFNYZPzJhEkeViG3VoElTvC2Mc0fcQboRv5IeFz3sRtzjVgQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
