-
PDF
- Split View
-
Views
-
Cite
Cite
Paul Visendi, Wanjiku Ng'ang'a, Wallace Bulimo, Richard Bishop, James Ochanda, Etienne P. de Villiers, TparvaDB: a database to support Theileria parva vaccine development, Database, Volume 2011, 2011, bar015, https://doi.org/10.1093/database/bar015
Close - Share Icon Share
Abstract
We describe the development of TparvaDB, a comprehensive resource to facilitate research towards development of an East Coast fever vaccine, by providing an integrated user-friendly database of all genome and related data currently available for Theileria parva. TparvaDB is based on the Generic Model Organism Database (GMOD) platform. It contains a complete reference genome sequence, Expressed Sequence Tags (ESTs), Massively Parallel Signature Sequencing (MPSS) expression tag data and related information from both public and private repositories. The Artemis annotation workbench provides online annotation functionality. TparvaDB represents a resource that will underpin and promote ongoing East Coast fever vaccine development and biological research.
Database URL:http://tparvadb.ilri.cgiar.org
Introduction
Theileria parva is a tick-transmitted haemoprotozoan parasite that causes an acute and often fatal disease of cattle, East Coast fever (ECF) (1). ECF severely constrains the livelihoods of poor livestock-keepers in sub-Saharan Africa. Current control methods include use of acaricides to limit tick populations, drug-treatment of cattle exhibiting clinical symptoms and deployment of a live vaccine that involves infection with a potentially lethal dose of cryo-preserved sporozoites and simultaneous treatment with long-acting oxy-tetracycline (2). A subunit vaccine will provide a long-term solution to this socio-economically important constraint to livestock development in Eastern and Southern Africa.
The completion of the genome sequence of T. parva Muguga (3) represents an important milestone in research on the parasite biology, and has contributed to the identification of candidate schizont antigens for vaccine development targeting this stage of the parasite (4). It is also an important resource for apicomplexan comparative genomics, in particular with Plasmodium falciparum, which causes malaria in humans and T. annulata (5), the cause of tropical bovine Theileriosis, a related disease of cattle that has a wide range in North Africa and South and East Asia. To date the utilization of the T. parva genome and associated information by interested scientists has been limited by the lack of a user-friendly interface that provides access not only to genome data but the large set of expression data from MPSS (6) and EST data for the schizont stage and additional unpublished and published microarray expression data (7). In addition, the current system of access does not easily allow for updating of annotation and curation as new data becomes available (8).
TparvaDB architecture
To address the need to provide a comprehensive resource to facilitate research in the development of an ECF vaccine and comparative Apicomplexan genomics, TparvaDB was developed to provide a unified platform for all genome and related T. parva data (Figure 1). TparvaDB was built using components of the Generic Model Organism Database (GMOD) system (http://www.gmod.org). The core component of TparvaDB is Chado, an ontology driven relational database schema (9), using the PostgreSQL open source database management system (http://www.postgresql.org). Ontologies used in TparvaDB include Sequence (10), Gene (11) and Relationship (12) ontologies.
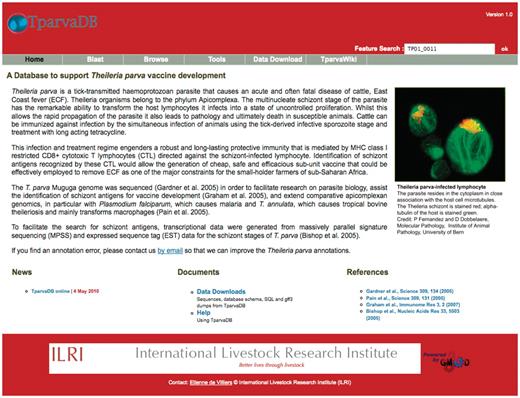
Screenshot of TparvaDB interface providing access to several online tools.
Theileria parva Muguga genome data in GenBank format was converted to GFF3 format and loaded into the TparvaDB Chado database using Perl scripts. EST, MPSS and SignalP data was similarly converted to GFF3 format and uploaded to the database. Several tools were included in TparvaDB to provide functionality. These include, GBrowse, NCBI Blast and an online annotation tool Artemis (13).
GBrowse, is a platform independent, extensible web-based graphical interface application for the visualization of genomic features stored in a database (14). In TparvaDB, GBrowse utilizes the Bio::DB::Das::Chado perl interface to connect directly to the TparvaDB Chado database, and the Bio::Graphics perl libraries to create images. GBrowse allows a simultaneous genome overview combined with the facility for a detailed view of specific regions of the genome. A user can query chromosomal regions of interest and visualise specific features such as annotated genes, ESTs and MPSS data mapped to a specific region (Figure 2). Online data analysis plug-ins provides detailed information on specific features that can be downloaded.
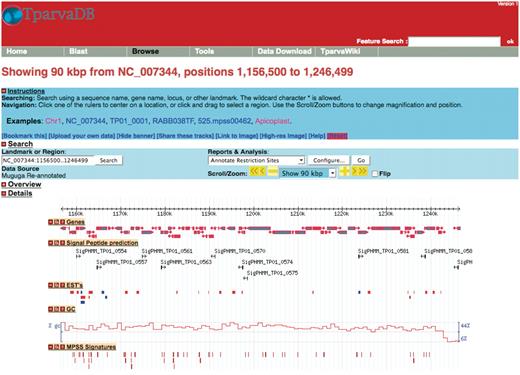
A representative example of the GBrowse interface. Tracks display information on annotated genes, predicted signal peptides, mapped ESTs, %GC content and MPSS signatures in a region of Chromosome 1.
The NCBI Blast (15) tool allows homology searches against either nucleotide or protein sequences from T. parva (Muguga). In future, database releases data for other T. parva isolates that are currently being sequenced, and will be included as they are completed.
We implemented an online annotation functionality using Artemis, a DNA sequence viewer and annotation tool (13). Artemis supports Chado databases through the use of the iBATIS DataMapper API (http://ibatis.apache.org), allowing real-time connection to the underlying TparvaDB Chado database. Users with granted permissions can login to the Chado database remotely and make changes to T. parva gene models online through the Artemis interface (Figure 3).
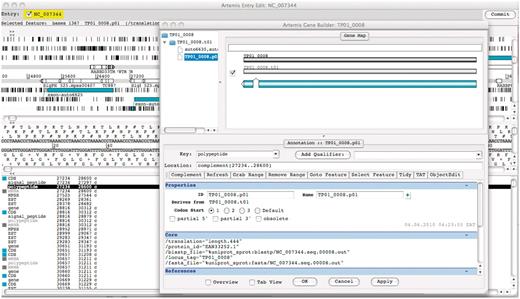
Screenshot of Artemis annotation tool allowing direct access to T. parva genome data in the TparvaDB database for online annotation. Background image depicts Artemis interface and foreground image depict Gene Builder view.
Theileria parva re-annotation
As mentioned several additional T. parva isolates are currently being sequenced which will enable comparative genomics analysis to aid ECF vaccine research. One of the most effective methods to annotate a newly sequenced genome is to compare it with a well-annotated closely related genome using computational tools and databases. In order to improve the quality of the reference genome, we re-annotated T. parva Muguga genome using data that had become available since the original annotation, most importantly a large EST data set generated since the first genome was sequenced. In this exercise, we used TparvaDB to re-annotate the T. parva Muguga genome using the new data. Theileria parva schizont and sporozoite EST data were downloaded from NCBI dbEST database (16), mapped to the T. parva genome using both PASA (17) and BLAT (18), and results loaded into TparvaDB. The original T. parva genome data and the mapped EST data were visualized and gene models manually annotated using Artemis. In total, 13 new gene models were created and 23 gene models were corrected as shown in Table 1.
| Chromosome . | Gene model . | Change . | Comment . |
|---|---|---|---|
| 1 | TP01_0115 | Modified CDS | |
| 1 | TP01_0289 | Modified CDS | Exon added |
| 1 | TP01_0869 | Modified CDS | Exon added |
| 1 | TP01_1252 | New CDS | |
| 1 | TP01_1253 | New CDS | |
| 1 | TP01_1254 | Modified CDS | Exon added |
| 1 | TP01_1255 | New CDS | |
| 2 | TP02_0127 | Modified CDS | Exon added |
| 2 | TP02_0140 | Modified CDS | Exon added |
| 2 | TP02_0173 | Modified CDS | Exon added |
| 2 | TP02_0181 | Modified CDS | Exon added |
| 2 | TP02_0209 | Modified CDS | Exon added |
| 2 | TP02_0236 | Modified CDS | Exon added |
| 2 | TP02_0247 | Modified CDS | Exon added |
| 2 | TP02_0942 | Modified CDS | Exon added |
| 2 | TP02_0980 | New CDS | |
| 2 | TP02_0981 | New CDS | |
| 2 | TP02_0982 | New CDS | |
| 2 | TP02_0983 | New CDS | |
| 2 | TP02_0984 | New CDS | |
| 2 | TP02_0986 | New CDS | |
| 3 | TP03_0029 | Modified CDS | Exon added |
| 3 | TP03_0041 | Modified CDS | Exon added |
| 3 | TP03_0224 | Modified CDS | Four exons added |
| 3 | TP03_0349 | Modified CDS | Exon added |
| 3 | TP03_0413 | Modified CDS | Exon added |
| 3 | TP03_0542 | Modified CDS | Exon added |
| 3 | TP03_0574 | Modified CDS | Exon added |
| 3 | TP03_0792 | Modified CDS | Exon added |
| 3 | TP03_0862 | Modified CDS | Exon added |
| 3 | TP03_0945 | New CDS | |
| 3 | TP03_0946 | New CDS | |
| 4 | TP04_0943 | New CDS | |
| 4 | TP04_0566 | Modified CDS | Exon added |
| 4 | TP04_0945 | New CDS | |
| 4 | TP04_0843 | Modified CDS | Exon added |
| Chromosome . | Gene model . | Change . | Comment . |
|---|---|---|---|
| 1 | TP01_0115 | Modified CDS | |
| 1 | TP01_0289 | Modified CDS | Exon added |
| 1 | TP01_0869 | Modified CDS | Exon added |
| 1 | TP01_1252 | New CDS | |
| 1 | TP01_1253 | New CDS | |
| 1 | TP01_1254 | Modified CDS | Exon added |
| 1 | TP01_1255 | New CDS | |
| 2 | TP02_0127 | Modified CDS | Exon added |
| 2 | TP02_0140 | Modified CDS | Exon added |
| 2 | TP02_0173 | Modified CDS | Exon added |
| 2 | TP02_0181 | Modified CDS | Exon added |
| 2 | TP02_0209 | Modified CDS | Exon added |
| 2 | TP02_0236 | Modified CDS | Exon added |
| 2 | TP02_0247 | Modified CDS | Exon added |
| 2 | TP02_0942 | Modified CDS | Exon added |
| 2 | TP02_0980 | New CDS | |
| 2 | TP02_0981 | New CDS | |
| 2 | TP02_0982 | New CDS | |
| 2 | TP02_0983 | New CDS | |
| 2 | TP02_0984 | New CDS | |
| 2 | TP02_0986 | New CDS | |
| 3 | TP03_0029 | Modified CDS | Exon added |
| 3 | TP03_0041 | Modified CDS | Exon added |
| 3 | TP03_0224 | Modified CDS | Four exons added |
| 3 | TP03_0349 | Modified CDS | Exon added |
| 3 | TP03_0413 | Modified CDS | Exon added |
| 3 | TP03_0542 | Modified CDS | Exon added |
| 3 | TP03_0574 | Modified CDS | Exon added |
| 3 | TP03_0792 | Modified CDS | Exon added |
| 3 | TP03_0862 | Modified CDS | Exon added |
| 3 | TP03_0945 | New CDS | |
| 3 | TP03_0946 | New CDS | |
| 4 | TP04_0943 | New CDS | |
| 4 | TP04_0566 | Modified CDS | Exon added |
| 4 | TP04_0945 | New CDS | |
| 4 | TP04_0843 | Modified CDS | Exon added |
| Chromosome . | Gene model . | Change . | Comment . |
|---|---|---|---|
| 1 | TP01_0115 | Modified CDS | |
| 1 | TP01_0289 | Modified CDS | Exon added |
| 1 | TP01_0869 | Modified CDS | Exon added |
| 1 | TP01_1252 | New CDS | |
| 1 | TP01_1253 | New CDS | |
| 1 | TP01_1254 | Modified CDS | Exon added |
| 1 | TP01_1255 | New CDS | |
| 2 | TP02_0127 | Modified CDS | Exon added |
| 2 | TP02_0140 | Modified CDS | Exon added |
| 2 | TP02_0173 | Modified CDS | Exon added |
| 2 | TP02_0181 | Modified CDS | Exon added |
| 2 | TP02_0209 | Modified CDS | Exon added |
| 2 | TP02_0236 | Modified CDS | Exon added |
| 2 | TP02_0247 | Modified CDS | Exon added |
| 2 | TP02_0942 | Modified CDS | Exon added |
| 2 | TP02_0980 | New CDS | |
| 2 | TP02_0981 | New CDS | |
| 2 | TP02_0982 | New CDS | |
| 2 | TP02_0983 | New CDS | |
| 2 | TP02_0984 | New CDS | |
| 2 | TP02_0986 | New CDS | |
| 3 | TP03_0029 | Modified CDS | Exon added |
| 3 | TP03_0041 | Modified CDS | Exon added |
| 3 | TP03_0224 | Modified CDS | Four exons added |
| 3 | TP03_0349 | Modified CDS | Exon added |
| 3 | TP03_0413 | Modified CDS | Exon added |
| 3 | TP03_0542 | Modified CDS | Exon added |
| 3 | TP03_0574 | Modified CDS | Exon added |
| 3 | TP03_0792 | Modified CDS | Exon added |
| 3 | TP03_0862 | Modified CDS | Exon added |
| 3 | TP03_0945 | New CDS | |
| 3 | TP03_0946 | New CDS | |
| 4 | TP04_0943 | New CDS | |
| 4 | TP04_0566 | Modified CDS | Exon added |
| 4 | TP04_0945 | New CDS | |
| 4 | TP04_0843 | Modified CDS | Exon added |
| Chromosome . | Gene model . | Change . | Comment . |
|---|---|---|---|
| 1 | TP01_0115 | Modified CDS | |
| 1 | TP01_0289 | Modified CDS | Exon added |
| 1 | TP01_0869 | Modified CDS | Exon added |
| 1 | TP01_1252 | New CDS | |
| 1 | TP01_1253 | New CDS | |
| 1 | TP01_1254 | Modified CDS | Exon added |
| 1 | TP01_1255 | New CDS | |
| 2 | TP02_0127 | Modified CDS | Exon added |
| 2 | TP02_0140 | Modified CDS | Exon added |
| 2 | TP02_0173 | Modified CDS | Exon added |
| 2 | TP02_0181 | Modified CDS | Exon added |
| 2 | TP02_0209 | Modified CDS | Exon added |
| 2 | TP02_0236 | Modified CDS | Exon added |
| 2 | TP02_0247 | Modified CDS | Exon added |
| 2 | TP02_0942 | Modified CDS | Exon added |
| 2 | TP02_0980 | New CDS | |
| 2 | TP02_0981 | New CDS | |
| 2 | TP02_0982 | New CDS | |
| 2 | TP02_0983 | New CDS | |
| 2 | TP02_0984 | New CDS | |
| 2 | TP02_0986 | New CDS | |
| 3 | TP03_0029 | Modified CDS | Exon added |
| 3 | TP03_0041 | Modified CDS | Exon added |
| 3 | TP03_0224 | Modified CDS | Four exons added |
| 3 | TP03_0349 | Modified CDS | Exon added |
| 3 | TP03_0413 | Modified CDS | Exon added |
| 3 | TP03_0542 | Modified CDS | Exon added |
| 3 | TP03_0574 | Modified CDS | Exon added |
| 3 | TP03_0792 | Modified CDS | Exon added |
| 3 | TP03_0862 | Modified CDS | Exon added |
| 3 | TP03_0945 | New CDS | |
| 3 | TP03_0946 | New CDS | |
| 4 | TP04_0943 | New CDS | |
| 4 | TP04_0566 | Modified CDS | Exon added |
| 4 | TP04_0945 | New CDS | |
| 4 | TP04_0843 | Modified CDS | Exon added |
Future development of T. parva DB
To expedite selection of potential vaccine targets, and to determine the impact and potential limitations of extensive vaccine deployment, additional T. parva genomes are being sequenced. These new genomes can be quickly annotated using TparvaDB as they become available. We plan to incorporate a comparative genomics component to TparvaDB to facilitate identification of conserved and divergent regions between isolates. The comparative genomics component could either be implemented through GBrowse_syn (http://gmod.org/wiki/GBrowse_syn), a GBrowse-based synteny browser or by interfacing TparvaDB with other comparative database systems such as Sybil (http://sybil.sourceforge.net/). In addition, the ECF vaccine development project is generating immunological data on additional candidate antigens, bovine class I MHC sequences and other immunological data that can also be incorporated in TparvaDB. To manage this increasingly sophisticated information more efficiently, we plan to implement a query system based on BioMart, a query-oriented data management system that provides ‘data mining’-like searches of complex descriptive data (19).
Funding
Multilateral core funding provided to the International Livestock Research Institute supported this work. This is ILRI publication number IL-201101. Funding for open access charge: International Livestock Research Institute.
Conflict of interest. None declared.



