-
PDF
- Split View
-
Views
-
Cite
Cite
Robin A. Haw, David Croft, Christina K. Yung, Nelson Ndegwa, Peter D'Eustachio, Henning Hermjakob, Lincoln D. Stein, The Reactome BioMart, Database, Volume 2011, 2011, bar031, https://doi.org/10.1093/database/bar031
Close - Share Icon Share
Abstract
Reactome is an open source, expert-authored, manually curated and peer-reviewed database of reactions, pathways and biological processes. We provide an intuitive web-based user interface to pathway knowledge and a suite of data analysis tools. The Reactome BioMart provides biologists and bioinformaticians with a single web interface for performing simple or elaborate queries of the Reactome database, aggregating data from different sources and providing an opportunity to integrate experimental and computational results with information relating to biological pathways.
Database URL:http://www.reactome.org
Project description
The Reactome project aims to systematically associate human proteins with their molecular and cellular functions in order to create a knowledgebase of human biological reactions, pathways and processes that can be used both as an online encyclopedia and as a systems biology platform for data mining and analysis (1–4). Reactome curators create these annotations in collaboration with domain experts who serve as authors and peer reviewers. The resulting molecular anatomies of pathways are extensively cross-referenced to the Ensembl, NCBI Entrez Gene and UniProt databases, the HapMap and UCSC Genome Browsers, the ChEBI and KEGG Compound small molecule databases, GO and PubMed (5–14).
As of June 2011 (Release 37), the Reactome database holds 6248 human proteins organized into 4354 reactions and 1153 pathways, and supported by 8942 publications. Examples of biological pathways in Reactome include signaling, innate and acquired immune function, transcriptional regulation, translation, apoptosis and classical intermediary metabolism (15, 16). The Reactome database includes computationally inferred pathways and reactions for twenty evolutionary divergent model organisms, including all 12 of the species in the GO Reference Genome annotation project (11).
Reactome embodies a reductionist data model, which represents diverse events in biology as reactions located in subcellular compartments that convert input physical entities into output physical entities. ‘Conversion’ encompasses not only the chemical transformations of classical biochemistry, but transport of molecules from one location to another, ligand–receptor binding in the context of signal transduction, and the modification and degradation of macromolecules. Reactome captures physical entities and events in Protégé, a knowledge-based framework (17). Classes (or frames) describe the different concepts such as reactions, physical entities and cellular compartments. Attributes (or slots) contain the properties of the instances such as the identities of the molecules that participate as inputs and outputs of a reaction. Physical entities of reactions can be proteins, nucleic acids, macromolecular complexes, chemical compounds or photons. All entities are located in subcellular compartments and macromolecular ones can also be cleaved, modified, or adopt different structural conformations. Each modified molecule in Reactome is represented as a separate entity, and the modification event can be annotated as a reaction wherein the input is the unmodified entity and the output is the altered form. Post-translational modifications are represented in Reactome with terms from PSI-MOD (18). With the annotation of post-translational modifications, conformational changes and subcellular locations, the number of variant instances of a physical entity can be large. The ‘reference physical entities’ class addresses this situation by storing the invariant features of a molecule such as its names, molecular structure and links to external reference databases such as EMBL for nucleic acids, UniProt for proteins and ChEBI for small molecules (9, 10, 19). Macromolecular complexes participate in numerous Reactome reactions and are formed by the association of two or more other entities (e.g. proteins, nucleic acids, small molecules and other complexes). The attributes capture the identities of components of the complex and its subcellular location. A glossary of class definitions and a full specification of the data model are available (http://wiki.reactome.org/index.php/Glossary_Data_Model and http://www.reactome.org/cgi-bin/classbrowser?DB=gk_current, respectively).
Accessible from the Reactome website, the Simple search tool allows Reactome users to query the entire Reactome database and website. Users can submit a word, database identifier or phrase and retrieve a list of corresponding database records. The Advanced (Extended) search provides customizable, logical and complex queries of the Reactome database. Specifically, this Extended search method delivers full schema-based queries for instances in the database by multiple attribute values. Queries can be combined together with boolean ‘AND’ operators. To support more systematic data mining, interactive analysis and modeling, Reactome offers pathway and reaction data in BioPAX, SBML, PSI-MITAB and Protégé formats, and as a MySQL database (17, 20, 21). A challenge for biologists and bioinformaticians who want to perform advanced integrative searches across multiple databases is that individual queries are time consuming and the results generated usually require further formatting. In this article, we focus on the simple-to-use and highly customizable Reactome BioMart interface, a platform for scientists to efficiently query and integrate pathway and other experimental datasets. Users of Reactome can, for example, find the Affymetrix probe identifiers associated with the genes in selected Reactome pathways by using BioMart to link a Reactome query to an ENSEMBL query through a single web interface.
Reactome BioMart
Developed jointly by the Ontario Institute for Cancer Research and European Bioinformatics Institute (EBI), BioMart (www.biomart.org) is a powerful query-oriented data management system (22–24). The BioMart portal provides an easy-to-use interface that allows the user to rapidly create simple or complex bulk queries of a database like Reactome without any specialist knowledge of its data model or programming skills. The user has control over both how the data is ‘filtered’, to limit the records that are integrated and the ‘attributes’, corresponding to columns of data that are included in the results. The existence of over 40 publicly accessible BioMart databases and the ability to combine two or more BioMart datasets in a single query permit the integration of biological information drawn from multiple sources in multiple original formats. The Reactome BioMart web interface is accessible from the ‘Tools’ menu located in the main navigation bar on the Reactome Home page (and most of the Reactome webpages). The Reactome BioMart is also accessible from the BioMart Central Portal at http://www.biomart.org/biomart/martview/, where other BioMarts are also available (25, 26). Simple or complex queries can be generated through the BioMart web interface to query the Reactome database. The preformatted queries can be accessed at the top of the BioMart page while the regular BioMart query interface is located below the canned query selector. Pathway data within Reactome BioMart can also be programmatically accessed using Perl API and URL/XML based queries. At the top of the Reactome BioMart web interface are buttons allowing users to see what their queries look like in the web services API query and the URL/XML format. The Reactome BioMart also makes available a SOAP web services API to allow users and third-party analysis workflows including Galaxy, BioConductor and Cytoscape, to access Reactome data. (27–29).
Query examples
Queries of Reactome data through BioMart proceed in three steps. Selecting the information source to search in Reactome initializes the query. User-selected filters are then applied to refine the search. Reactome BioMart supports numerous internal and external database accession numbers and identifiers, and batch querying to limit the query. Finally, particular characteristics of the filtered data are selected with drop-down menus or radio buttons in the user interface, and displayed through the output webpage. Reactome provides three types of BioMart queries: preformatted (canned), regular and federated. A set of standard (or preformatted) queries can be used without detailed knowledge of the BioMart query interface (Figure 1). Data entry will differ, depending on whether single or multiple data items are permitted. When a single item is allowed, the entry form presents a selector to choose the item, e.g. species. If multiple data items are allowed, the entry form presents a text box, in which to enter the items separated by newlines, e.g. a list of Entrez Gene identifiers. The standard query selector allows the user to choose from one of the currently available queries, to:
Find list of pathways for specific species (multiple data items). The user can use this query to list all pathways known to Reactome for a species of choice.
Find list of reactions for specific pathways (multiple data items). Given a list of Reactome stable pathway identifiers, this canned query retrieves all of the reactions involved in the pathways. All reactions involved in all known pathways will be retrieved if the query is initiated without any data values.
Find list of proteins for specific pathways (multiple data items). Given a list of Reactome stable pathway identifiers, this canned query retrieves all of the proteins involved in the pathways. If this query is initiated without any data values, all proteins involved in all known pathways will be returned.
Find list of complexes for specific proteins. This canned query will find all of the complexes in Reactome whose components include any of a submitted list of protein UniProt identifiers. It will return all complexes and their associated proteins, if no data values were submitted in the original query. An example of this canned query is shown in Table 1.
Find list of pathways for specific genes. Given a list of Entrez gene identifiers, this canned query retrieves all of the pathways in Reactome involving those genes. All pathways and their associated genes will be returned if the query is initiated without submitting any data values.
Find list of genes for specific pathways. Given a list of Reactome stable pathway identifiers, this canned query retrieves all of the genes whose protein products are involved in the pathways. If the user initiated this query without submitting any data values, all genes involved in all known pathways will be returned.
Find list of reactions for specific genes. Given a list of Entrez Gene IDs, this canned query retrieves all of the reactions in Reactome involving the protein products of those genes. All reactions and their associated genes will be returned if the query is initiated without submitting any data values.
By default, the query will return a preview of the results encompassing the first ten rows of data. Once the user has reviewed the data, possibly making modifications to the original query, the full data set can be exported. The results can be downloaded in a variety of formats such as a HTML table, tab and comma separated values files, or an Excel spreadsheet. For large and complex queries, users have an additional option to download a compressed results file (.gz) or to be notified by email when the file is ready for download.
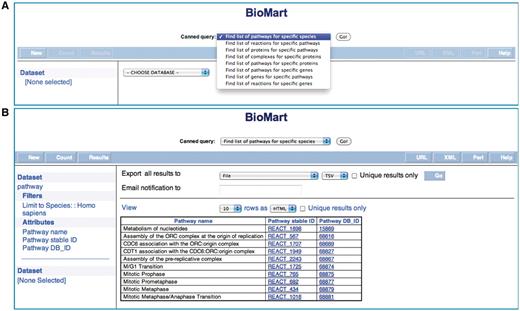
Reactome BioMart Canned Query. (A) The canned query selector allows the user to choose from one of the currently available queries. (B) The results table for the canned query.
| Datasets . | Filters . | Attributes . |
|---|---|---|
| complex | Limit to complexes containing these IDs: REACT_4500 | Complex Species name |
| Protein DB_ID | ||
| Protein Identifier | ||
| Complex DB_ID | ||
| Complex stable ID | ||
| Protein name |
| Datasets . | Filters . | Attributes . |
|---|---|---|
| complex | Limit to complexes containing these IDs: REACT_4500 | Complex Species name |
| Protein DB_ID | ||
| Protein Identifier | ||
| Complex DB_ID | ||
| Complex stable ID | ||
| Protein name |
Reactome dataset ‘filters’ and ‘attributes’ required to search and extract human protein and reaction annotations.
| Datasets . | Filters . | Attributes . |
|---|---|---|
| complex | Limit to complexes containing these IDs: REACT_4500 | Complex Species name |
| Protein DB_ID | ||
| Protein Identifier | ||
| Complex DB_ID | ||
| Complex stable ID | ||
| Protein name |
| Datasets . | Filters . | Attributes . |
|---|---|---|
| complex | Limit to complexes containing these IDs: REACT_4500 | Complex Species name |
| Protein DB_ID | ||
| Protein Identifier | ||
| Complex DB_ID | ||
| Complex stable ID | ||
| Protein name |
Reactome dataset ‘filters’ and ‘attributes’ required to search and extract human protein and reaction annotations.
The regular Reactome BioMart query interface enables the user to define simple or complex queries (Figure 2). The first step is selecting the ‘database’ and ‘dataset’ to initiate the query. Reactome provides four datasets that are accessible to the BioMart query, ‘complex’, ‘pathway’, ‘interaction’ and ‘reaction’. For example, selecting the ‘reaction’ dataset will restrict the query to include reaction annotations. The next step is to select the ‘Filters’ to restrict the query, e.g. ‘Limit to Species’—Homo sapiens, to retrieve data for H. sapiens rather than all species known to Reactome. Selecting ‘Attributes’ will specifically define what data is displayed in the results as shown in the example in Table 2. As with the canned queries, the results of the regular BioMart query are previewed as an HTML table or can be downloaded in the supported formats.
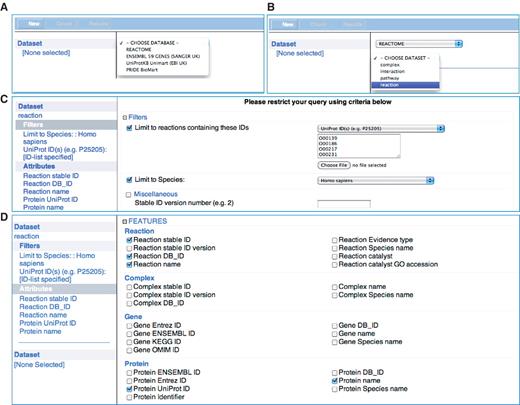
Reactome BioMart Regular Query. (A) The ‘database’ selector selects the REACTOME database. (B) The ‘dataset’ drop-down menu. (C) The ‘filters’ page that allows the user to narrow down the query to the UniProt identifiers provided and human annotations. (D) The reaction ‘attributes’ that determines the columns to be displayed in the results table.
| Datasets . | Filters . | Attributes . |
|---|---|---|
| reaction | Limit to Species: Homo sapiens | Reaction stable ID |
| Reaction DB_ID | ||
| Reaction name | ||
| Protein UniProt ID | ||
| Protein name |
| Datasets . | Filters . | Attributes . |
|---|---|---|
| reaction | Limit to Species: Homo sapiens | Reaction stable ID |
| Reaction DB_ID | ||
| Reaction name | ||
| Protein UniProt ID | ||
| Protein name |
Reactome dataset ‘filters’ and ‘attributes’ required to search and extract human protein and reaction annotations.
| Datasets . | Filters . | Attributes . |
|---|---|---|
| reaction | Limit to Species: Homo sapiens | Reaction stable ID |
| Reaction DB_ID | ||
| Reaction name | ||
| Protein UniProt ID | ||
| Protein name |
| Datasets . | Filters . | Attributes . |
|---|---|---|
| reaction | Limit to Species: Homo sapiens | Reaction stable ID |
| Reaction DB_ID | ||
| Reaction name | ||
| Protein UniProt ID | ||
| Protein name |
Reactome dataset ‘filters’ and ‘attributes’ required to search and extract human protein and reaction annotations.
Reactome BioMart facilitates query integration across different datasets, providing the option of combining annotation data from diverse sources. The second ‘Dataset’ link in the left hand panel is used to choose another dataset, enabling the integration of Reactome data with a dataset from another database (Figure 3). To formulate a federated query, the datasets involved need to share at least one common attribute, typically a molecule identifier. For example, the ENSEMBL identifier provides the data linkage to create a query to combine the Reactome ‘pathways’ dataset with an ENSEMBL dataset. Currently, it is possible to query Reactome with ENSEMBL and UniProt (7, 9) directly from the Reactome BioMart Portal. Through the Central BioMart Portal, other datasets can be merged with a Reactome dataset query, such as PRIDE, COSMIC, International Knockout Mouse Consortium (IKMC) Projects (Table 3), Vectorbase and Wellcome Trust Sanger Institute (WTSI) Mouse Genetics Project (30–35).
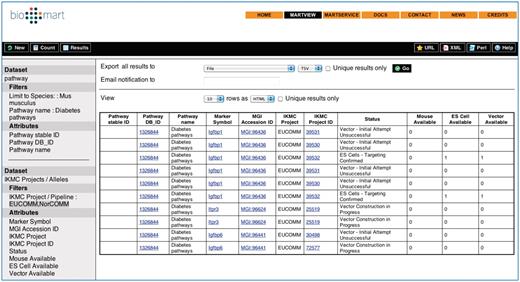
Combined Reactome-IKMC Query results from the BioMart Central Portal. The Reactome and IKMC dataset ‘filters’ and ‘attributes’ are visible on the left of the results table.
| Datasets . | Filters . | Attributes . |
|---|---|---|
| pathway | Limit to Species: | Pathway stable ID |
| (Reactome) | Mus musculus | Pathway DB_ID |
| Pathway name: Diabetes pathways | Pathway name | |
| IKMC GENES AND PRODUCTS (IKMC) | IKMC Project/Pipeline: EUCOMM and NorCOMM | Marker Symbol |
| MGI Accession ID | ||
| IKMC Project | ||
| IKMC Project ID | ||
| Status | ||
| Mouse Available | ||
| ES Cell Available | ||
| Vector Available |
| Datasets . | Filters . | Attributes . |
|---|---|---|
| pathway | Limit to Species: | Pathway stable ID |
| (Reactome) | Mus musculus | Pathway DB_ID |
| Pathway name: Diabetes pathways | Pathway name | |
| IKMC GENES AND PRODUCTS (IKMC) | IKMC Project/Pipeline: EUCOMM and NorCOMM | Marker Symbol |
| MGI Accession ID | ||
| IKMC Project | ||
| IKMC Project ID | ||
| Status | ||
| Mouse Available | ||
| ES Cell Available | ||
| Vector Available |
Dataset ‘filters’ and ‘attributes’ required to search and extract mouse Diabetes pathway annotations from Reactome and information about the availability of mouse knockout reagents from the IKMC Project.
| Datasets . | Filters . | Attributes . |
|---|---|---|
| pathway | Limit to Species: | Pathway stable ID |
| (Reactome) | Mus musculus | Pathway DB_ID |
| Pathway name: Diabetes pathways | Pathway name | |
| IKMC GENES AND PRODUCTS (IKMC) | IKMC Project/Pipeline: EUCOMM and NorCOMM | Marker Symbol |
| MGI Accession ID | ||
| IKMC Project | ||
| IKMC Project ID | ||
| Status | ||
| Mouse Available | ||
| ES Cell Available | ||
| Vector Available |
| Datasets . | Filters . | Attributes . |
|---|---|---|
| pathway | Limit to Species: | Pathway stable ID |
| (Reactome) | Mus musculus | Pathway DB_ID |
| Pathway name: Diabetes pathways | Pathway name | |
| IKMC GENES AND PRODUCTS (IKMC) | IKMC Project/Pipeline: EUCOMM and NorCOMM | Marker Symbol |
| MGI Accession ID | ||
| IKMC Project | ||
| IKMC Project ID | ||
| Status | ||
| Mouse Available | ||
| ES Cell Available | ||
| Vector Available |
Dataset ‘filters’ and ‘attributes’ required to search and extract mouse Diabetes pathway annotations from Reactome and information about the availability of mouse knockout reagents from the IKMC Project.
Discussion
Reactome is an online, manually curated pathway resource that provides an integrated view of the molecular details of biological processes that range from metabolism to DNA replication and repair to signaling cascades. Its data model allows these diverse processes to be represented in a consistent way to facilitate usage as online text and as a resource for data mining, modeling and analysis of large-scale expression datasets. The Reactome BioMart web interface allows both biologists and bioinformaticians to easily query and retrieve Reactome pathway, reaction, complex and interaction annotations and to integrate this information with their own experimental data. Our curation practice and data model allow Reactome to capture pathway annotations encompassing a very broad range of human biology. As we extend Reactome annotations to new signaling pathways, tissue-specific processes and pathways including normal development as well as disease processes such as infection and malignant transformation, the content within the Reactome BioMart will expand to support these additional annotations. We have developed a new dataset in Reactome BioMart called ‘Protein’ (36). Once this dataset is released, it will enable the user to search and retrieve post-translational modification data for a protein such as the type of the modification, the modified residue, the coordinate of the modified residue on the protein sequence, the start and stop positions of the protein sequence, and the cellular compartment of the modified protein. The future integration of federated clinical datasets with Reactome BioMart will see the ability to search and integrate genomic, transcriptomic and epigenomic data with Reactome pathway data. For example, Reactome pathway data is available through The International Cancer Genome Consortium (ICGC) Data Portal that employs BioMart to provide access to data from 50 different tumor types and subtypes (37, 38). The Reactome group will continue to support the development and distribution of open software for the management of pathway information in order to encourage data standards, analysis and integration.
Funding
Development of the Reactome database was supported by grants from the National Human Genome Research Institute at the National Institutes of Health (grant number P41 HG003751); the European Union 6th Framework Programme ‘ENFIN’ (grant number LSHG-CT-2005-518254). Funding for open access charge: National Institutes of Health grant number P41 HG003751.
Conflict of interest. None declared.
Acknowledgements
Development of the Reactome website, data model and data analysis tools described in this article is a result of concerted work of the Reactome curators and developers. The authors are also grateful to the many scientists who collaborated with us to build the Reactome pathway content.



