-
PDF
- Split View
-
Views
-
Cite
Cite
Guy Tsafnat, Joseph Copty, Sally R. Partridge, RAC: Repository of Antibiotic resistance Cassettes, Database, Volume 2011, 2011, bar054, https://doi.org/10.1093/database/bar054
Close - Share Icon Share
Abstract
Antibiotic resistance in bacteria is often due to acquisition of resistance genes associated with different mobile genetic elements. In Gram-negative bacteria, many resistance genes are found as part of small mobile genetic elements called gene cassettes, generally found integrated into larger elements called integrons. Integrons carrying antibiotic resistance gene cassettes are often associated with mobile elements and here are designated ‘mobile resistance integrons’ (MRIs). More than one cassette can be inserted in the same integron to create arrays that contribute to the spread of multi-resistance. In many sequences in databases such as GenBank, only the genes within cassettes, rather than whole cassettes, are annotated and the same gene/cassette may be given different names in different entries, hampering analysis. We have developed the Repository of Antibiotic resistance Cassettes (RAC) website to provide an archive of gene cassettes that includes alternative gene names from multiple nomenclature systems and allows the community to contribute new cassettes. RAC also offers an additional function that allows users to submit sequences containing cassettes or arrays for annotation using the automatic annotation system Attacca. Attacca recognizes features (gene cassettes, integron regions) and identifies cassette arrays as patterns of features and can also distinguish minor cassette variants that may encode different resistance phenotypes (aacA4 cassettes and bla cassettes-encoding β-lactamases). Gaps in annotations are manually reviewed and those found to correspond to novel cassettes are assigned unique names. While there are other websites dedicated to integrons or antibiotic resistance genes, none includes a complete list of antibiotic resistance gene cassettes in MRI or offers consistent annotation and appropriate naming of all of these cassettes in submitted sequences. RAC thus provides a unique resource for researchers, which should reduce confusion and improve the quality of annotations of gene cassettes in integrons associated with antibiotic resistance.
Database URL:http://www2.chi.unsw.edu.au/rac.
Introduction
Antibiotic resistance in bacteria is often due to the acquisition of mobile resistance genes by horizontal (also called lateral) gene transfer (1) mediated by the actions of two different types of mobile elements. Some elements (e.g. plasmids) are able to move between cells, including those of different species, whereas others (e.g. transposons, insertion sequences) can move between DNA molecules in the same cell. In Gram-negative bacteria, in particular, a wide range of antibiotic resistance genes are found as part of small mobile elements of the second type, called gene cassettes (2). A gene cassette consists of a single, usually promoter-less, gene (or occasionally two) and a recombination site (attC, previously known as a 59-base element). The attC sites of different cassettes vary in length and sequence, but share conserved regions at their ends.
Gene cassettes can exist transiently as circles, but do not carry the machinery for their own movement and are usually found in a linear form integrated into larger elements called integrons (3). Integrons are defined by an intI gene and an attI recombination site and also include a Pc promoter (Figure 1A). The IntI integrase catalyses recombination between attI and attC (preferred over attC×attC) to insert a cassette into the integron, where the gene it carries can be expressed from Pc (3). Recombination occurs at a specific site so that in a linear, integrated cassette 7 bp of the attC site is found at the start of the cassette and the remainder is located beyond the cassette gene (Figure 1A). Several cassettes can be inserted in tandem in the same integron to create cassette arrays, which can contribute to simultaneous resistance to multiple antibiotics.
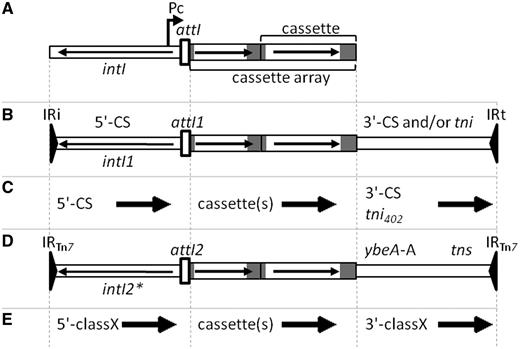
(A) General structure of an integron with intI, attI, Pc and a cassette array with two gene cassettes. Horizontal arrows indicate the extent and direction of genes. Grey regions represent attC sites. (B) General structure of Class 1 MRI, which may include the 3'-CS and part of the tni region of Tn402 or a complete tni402 region. (C) Names used by RAC to indicate conserved regions flanking cassette arrays in the two types of Class 1 MRI. Arrows define the forward direction used in the RAC annotations, which in the case of the 5'-CS is opposite to that of intI1. (D) Structure of a Class 2 integron associated with Tn7. The intI2 gene is usually truncated, indicated by an asterisk. ybeA-A is a cassette remnant with a truncated attC site and is followed by the tns transposition genes of Tn7. (E) Names used by RAC to indicate regions flanking cassette arrays in a class X MRI, where X is 2–5.
The first integrons were discovered due to their association with antibiotic resistance genes (4) then identification of other intI/attI combinations led to definition of different integron classes, with the original type becoming Class 1. Capture of intI1/attI1 (probably from a chromosomal location) by a transposon (5) gave a structure related to Tn402, with a complete transposition (tni) region and bounded by 25 bp inverted repeats designated IRi (integrase end) and IRt (tni end; Figure 1B). The most frequently observed Class 1 integrons in clinical isolates are derivatives of this structure in which only part of the tni region is present, preventing self-transposition. In these structures, a specific sequence called the 3′-conserved segment (3′-CS) is found beyond the cassette array and, similarly, the region between IRi and the start of the first cassette is termed the 5′-CS (Figure 1B). The current ‘functional’ definition of an integron refers to only the minimal integron components intI and attI but the original ‘structural’ definition of a Class 1 integron, which includes all of the Tn402-like transposon from IRi to IRt, is useful when annotating sequences, as it indicates the extent of the region that can potentially be transposed (6).
Although Class 1 integrons are the most commonly identified in clinical isolates, several other intI/attI combinations are also with associated mobile elements and resistance cassettes. intI2/attI2 of Class 2 integrons are usually found as part of the transposon Tn7 (7) in which conserved regions different from the 5′-CS and 3′-CS flank the cassette array (Figure 1D). intI3/attI3 of Class 3 integrons are thought to be associated with a transposon (8) and a few examples of Class 4 integrons have now been identified to be inserted in integrative conjugative elements (9), whereas the sequence of the only available example of a Class 5 integron, found on a plasmid (GenBank accession AJ277063), ends within the cassette array. Here, these five types of integron associated with mobile elements and carrying a few cassettes mainly encoding antibiotic resistance genes are designated ‘mobile-resistance integrons’ (MRIs) to distinguish them from chromosomal integrons (CIs; also referred to as ‘superintegrons’). CI have tens to hundreds of gene cassettes, mostly carrying genes of unknown function and typically with closely related attC sites, and are found on the chromosomes of many bacterial species (3).
The development of PCR methods to amplify cassette arrays using primers in the 5′-CS and 3′-CS of Class 1 integrons and the equivalent regions of other MRI and the increase in complete sequencing of resistance plasmids have led to an explosion in the number of cassette array sequences in databases such as GenBank. Unfortunately, the use of different nomenclature systems for antibiotic resistance genes, after which the gene cassettes are named, and a lack of a central archive for assigning these names have led to incorrect or inaccurate annotations in many available sequences. Often the same cassette is given different names in different sequences or different cassettes are given the same name, leading several authors to comment on the need to address this problem (10–12). In addition, most automated annotation systems focus on identifying open reading frames and the functions of the proteins that they encode. For gene cassettes, which may have been identified many times and which have known functions, the challenge is to name genes/cassettes consistently and to accurately identify the cassette boundaries.
Whereas several existing websites (listed below) provide information about antibiotic resistance genes, gene cassettes and/or integrons, none of them has been set up to include a complete list of known gene cassettes in MRI, to accurately and consistently annotate all resistance gene cassettes in MRI or to assign names to novel cassettes.
XXR (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=xxr) (13) is a program, which uses heuristics to predict attC sites. Whereas this can help to identify cassette boundaries, XXR does not attempt to identify or name cassette genes.
ACID (http://integron.biochem.dal.ca/ACID/phpbb3/) (14) identifies intI-like genes, attC sites and open reading frames in cassettes in publically available sequences, with manual editing to improve accuracy, and allows users to integrate their own data. The ACID algorithms were primarily aimed at annotating attC sites in CI and some of the more variable attC sites found in cassettes in MRI are currently missed. Cassette genes and cassettes are also assigned numbers that are unique to this database.
INTEGRALL (http://integrall.bio.ua.pt/) (15) includes a searchable list of available integron sequences that indicates which intI gene and which cassette array each contains. Whereas the spans and sequences of the intI and cassette genes can be accessed from the accession number, the positions of the gene cassettes themselves are not given and the gene names come from the GenBank entries and so suffer from the same inconsistencies.
The Antibiotic Resistance Database (ARDB; http://ardb.cbcb.umd.edu) (16) lists antibiotic resistance genes grouped by resistance functions and enables various searches against this database. Whereas this could be used in identification of cassette-borne genes, ARDB does not indicate which genes are found in gene cassettes and does not always use accepted antibiotic resistance gene nomenclature.
The Comprehensive Antibiotic Resistance Database (CARD; http://arpcard.mcmaster.ca) is an ontology of antibiotic resistance genes but currently does not indicate which resistance genes are cassette-borne. Searches will flag genes associated with antibiotic resistance, but do not identify gene cassettes.
We have developed the Repository of Antibiotic resistance Cassettes (RAC) to provide a unique solution to the problem of consistently annotating gene cassettes in MRI. RAC uniquely offers the following primary functions:
a central free repository of known cassettes conferring antibiotic resistance, using standard nomenclature systems where these have been established and listing alternative names where appropriate.
accurate and consistent annotation of gene cassettes in DNA sequences containing cassette arrays using the nomenclature systems defined in the repository.
a process for assignment of unique names for newly sequenced antibiotic resistance cassettes in MRI consistent with existing nomenclature systems and for adding new cassettes to the repository.
In this article, we describe RAC and the annotation and review processes it implements, and how these provide an easy-to-use solution for annotation of gene cassette arrays in MRI.
Browsing the cassette repository
The cassette repository was compiled from our previous review (2) and new cassettes identified since this was published (currently >40) and is freely accessible, without user registration, from the RAC homepage. RAC will continue to be updated with newly identified cassettes, including those submitted by users, and offers a central location for the community to share knowledge. RAC aims to include/annotate all cassettes found in MRI, whether or not they carry a resistance gene. Gene cassettes in CI will generally only be included if they have also been found in MRI or if they are closely related to known antibiotic resistance cassettes.
Clicking on the ‘Browse cassettes’ tab gives access to the entire cassette database, found under ‘All’. Nine sublists group cassettes conferring resistance to major classes of antibiotics (aminoglycosides, β-lactams, chloramphenicol, fosfomycin, macrolides, quinolones, rifampicin, trimethoprim), and those encoding small efflux proteins that give resistance to disinfectants. Some MRI include gene cassettes for which a function cannot be predicted (previously designated orfA, orf1 etc) and these are grouped under ‘gcu’ (for gene cassettes of unknown function) (2). The remaining cassettes found in MRI, including those with putative functions other than antibiotic resistance and the single known example of a cassette giving streptothricin resistance, are grouped under ‘Other’.
Each entry includes a unique cassette name, gives a GenBank accession number containing an exemplar sequence, where available, and the start and end positions of the cassette in this sequence. The exemplar chosen is generally the first reported complete cassette, unless possible sequence errors or other problems have been identified, but the annotations in that GenBank entry may not be correct. The cassette names also provide direct access to the exemplar cassette sequence. ‘Notes’ include information such as alternative cassette names and indicate where only a partial cassette sequence is available in GenBank.
Cassettes that are >98% identical are generally included under a single name, but cassette variants with minor sequence differences known to affect the resistance phenotype conferred are distinguished.
Variants of the aacA4 [also called aac(6')-Ib] cassette associated with resistance to either gentamicin (C at position 329, encoding serine) or amikacin (329T, encoding leucine) (17) are called aacA4-C329 and aacA4-T329. Two versions of the aacA4 cassette conferring low-level resistance to fluoroquinolones (18) with T283A G514T or T283C G514T mutations are distinguished as aacA4-crA and aacA4-crC, respectively.
A number of different families of β-lactamases have been identified (e.g. OXA-10-like, OXA-2-like, IMP, VIM, GES, VEB). Within these families a single amino acid difference, which may result in significant changes in activity, is sufficient for assignment of a new protein number. RAC distinguishes bla gene cassettes encoding different variants of each of the main β-lactamase types and the family to which they belong is indicated in the ‘Notes’.
If other cassette variants conferring distinct phenotypes are identified in the future, these will also be incorporated into the RAC database.
The annotation system—Attacca
We have previously reported an algorithm for an automatic annotation system (called Attacca) for bacterial DNA (19). Attacca follows a computational linguistic method (20–22), which consists of two parts: a lexical recognizer that identifies occurrences of features from a ‘feature database’ (FDB) in a sequence, and a parser that identifies larger-than-gene structures as patterns of such features. The RAC annotation service uses Attacca to annotate gene cassettes and the conserved regions of Classes 1–3 integrons flanking cassette arrays, the intI14 region and a putative 3′-flanking region in Class 4 integrons and the intI5 region in Class 5 integrons to identify cassette arrays in MRI.
The lexical recognizer uses BLASTn (23) to identify occurrences of any feature from the FDB in a sequence. A minimum identity with the FDB exemplar, as specified in each entry in the FDB (usually ≥98%), must be met. Any additional selection criteria will then be applied before a region is annotated as the appropriate feature. For example, the constraint ‘AT 329 HAS ‘c’ ’ must be met for annotation of an aacA4 cassette as the aacA4329-C variant. In the case of bla genes, the amino acid sequence translated from the cassette gene must be 100% identical to that of a specific variant in the FDB for annotation as that variant. If multiple gene cassettes satisfy the criteria for the same region, the one with the highest BLAST score is used as the annotation.
If after all cassettes have been identified, the sequence still contains regions of at least 25 bp that have not been annotated, Attacca uses BLASTn to search the feature database with these regions. This step identifies ‘partial’ copies of gene cassettes and other features. Annotation of partial gene cassettes is subject to the same criteria as complete cassettes, but as distinguishing nucleotides may be missing, identification is potentially less accurate.
Annotating sequences with RAC
RAC provides private workspaces for users to upload, edit and annotate nucleotide sequences containing gene cassettes through the ‘My Sequences’ tab. Registration using a valid e-mail address is required to access this feature, but is free for non-commercial use.
The ‘Annotate new sequence’ tab allows DNA sequences to be uploaded and submitted for annotation by Attacca. Each sequence submitted needs to be given a short description (used for identification) and the sequence type must be indicated e.g. obtained by cassette array PCR or as part of a longer sequence such as a whole plasmid (this information is used in deciding whether the annotation requires manual review). Adding information about whether the DNA was from a plasmid or the chromosome and the species it was obtained from is optional.
Sequences submitted to Attacca through RAC initially appear in the ‘My sequences’ as being ‘in progress’ (Figure 2). The annotation process typically takes <2 min but may be delayed for several hours depending on system load. When a sequence has been annotated, its status will change to ‘annotated’ and the annotation can be accessed through the View icon.
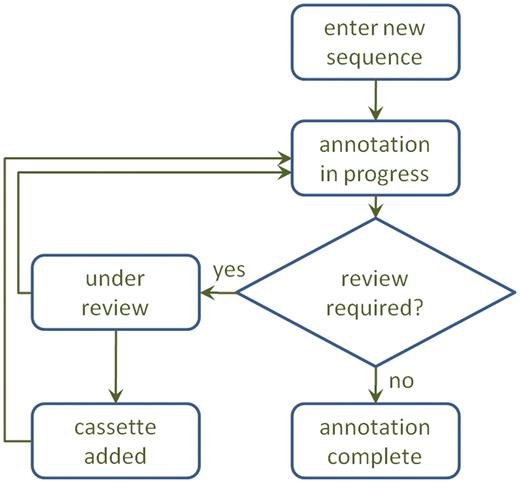
A diagram of the annotation process. After annotation, a sequence may be sent for review, which may result in a manual adjustment to the annotation and/or inclusion of a new cassette in the database.
The annotation provides names, spans and directions of regions identified as particular cassettes, as well as notes about the identified cassettes including e.g. alternative names by which they may be known (Figure 3). The 5′-CS and 3′-CS or tni region of Class 1 integrons and the equivalent regions marking the ends of cassette arrays in other classes of MRI (Figure 1C and E) are also annotated. Partial copies of features, created because the sequence ends within the feature or because of truncation by another feature, are indicated by hash symbol against the feature name. Insertion sequences that have already been identified as part of cassette arrays in MRI and other ‘non-cassette insertions’ are included in the FDB and will be annotated. Regions not included in the FDB and thus not identified by Attacca are marked by dashes.
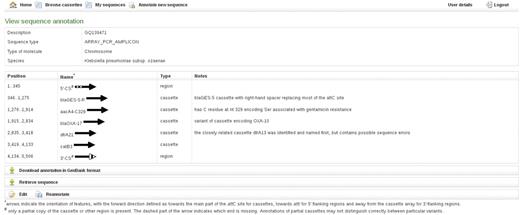
RAC annotation of an exemplar cassette array, including examples of partial features, a bla variant cassette, an aacA4 cassette and the type of additional information given in Notes.
The direction of each feature in the submitted sequence is indicated by an arrow. The forward direction of cassettes is defined as towards the main part of the attC site (Figure 1). The forward direction of the 5′ conserved flanking region is defined as towards attI (i.e. the opposite direction to the intI gene), whereas the forward direction of the 3′ conserved flanking region is defined as away from the cassette array (Figure 1). In the case of partial features, dashed part(s) of the arrows indicate whether the feature is truncated at the start, the end or both (Figure 3).
Manual review of annotations
Certain annotation results will cause the sequence to be sent to reviewers at the Centre for Infectious Diseases and Microbiology, University of Sydney for further examination. RAC will automatically redirect a sequence to review if:
the Attacca discovery heuristics (19) identify a gap in a cassette array that could correspond to a novel cassette;
a cassette encoding a potentially novel β-lactamase variant is detected; or
the type of sequence submitted (e.g. isolated cassette) suggests that a gene cassette should be present but a gene cassette is not found by Attacca.
In such cases, sequences will appear as ‘under review’ in ‘My Sequences’ until the manual review process is complete. The reviewers may contact the submitting user by e-mail if further details are necessary for the review (Figure 2).
Inclusion of new cassettes in the RAC database
An important aspect of RAC is to keep up to date by including new entries submitted by the research community. If a novel cassette is identified by a reviewer, they will e-mail the submitter suggesting a name based on the most similar cassette family and the next available number. Cassettes that are >98% identical to a cassette already in the repository will, in most cases, be annotated as this cassette and will not be included as separate features, unless a change in the associated phenotype is demonstrated. In the case of a cassette carrying a β-lactamase gene encoding a novel protein sequence, the user will be referred to the β-lactamase nomenclature website (www.lahey.org/Studies/) to obtain a unique name before the new cassette is registered with RAC.
Initially, the unique name given to a novel cassette will be reserved but the cassette will not be listed in RAC. The sequence will be available only to the submitting user and will only be included in annotations of other sequences from that user. If the user provides explicit written permission or when the cassette is published elsewhere (in GenBank or a journal), it will be added to the public cassette database with a reference to the appropriate GenBank entry or paper.
Limitations
RAC will only annotate features that are in the FDB, which presently includes gene cassettes, flanking regions found in MRI and selected insertion sequences and other regions currently known to interrupt cassette arrays. Additional features that are necessary for improving annotations will be added as they are identified. Attacca may not be able to properly annotate gene cassettes or other features in raw sequence data that contains many errors. For best results, users should manually check automatic base calls in chromatograms prior to annotation in RAC.
Conclusions
The RAC is an online knowledge base for microbiologists studying antibiotic resistance. It also provides convenient access to the Attacca automatic annotation engine that allows users to easily and accurately annotate cassette arrays in bacterial DNA sequences. Researchers using RAC can also contribute new gene cassettes to the knowledge base, obtain a unique and consistent name and share their sequences with other researchers.
Funding
Australian National Data Service, Data Capture (grant #DC5F), a New South Wales Health Capacity Building Infrastructure grants to the Australian Institute for Health Innovation (to G.T.); Australian National Health and Medical Research Council (grant 512396 to S.R.P.). Funding for open access charge: Centre for Health Informatics.
Conflict of interest. None declared.
Acknowledgements
We would like to thank Andrew Ginn, Agnieszka Wiklendt, Thu Betteridge, Piklu Roy Chowdhury and Lee Thomas for help with testing RAC. This work was presented as Poster P1816 at the 21st ECCMID/27th ICC. Milan, Italy, 7–10 May 2011.



