-
PDF
- Split View
-
Views
-
Cite
Cite
Chia-Jung Yang, Jung-Hsien Chiang, Gene ontology concept recognition using named concept: understanding the various presentations of the gene functions in biomedical literature, Database, Volume 2018, 2018, bay115, https://doi.org/10.1093/database/bay115
Close - Share Icon Share
Abstract
Objective: A major challenge in precision medicine is the development of patient-specific genetic biomarkers or drug targets. The firsthand information of the genes associated with the pathologic pathways of interest is buried in the ocean of biomedical literature. Gene ontology concept recognition (GOCR) is a biomedical natural language processing task used to extract and normalize the mentions of gene ontology (GO), the controlled vocabulary for gene functions across many species, from biomedical text. The previous GOCR systems, using either rule-based or machine-learning methods, treated GO concepts as separate terms and did not have an efficient way of sharing the common synonyms among the concepts.
Materials and Methods: We used the CRAFT corpus in this study. Targeting the compositional structure of the GO, we introduced named concept, the basic conceptual unit which has a conserved name and is used in other complex concepts. Using the named concepts, we separated the GOCR task into dictionary-matching and machine-learning steps. By harvesting the surface names used in the training data, we wildly boosted the synonyms of GO concepts via the connection of the named concepts and then enhanced the capability to recognize more GO concepts in the text. The source code is available athttps://github.com/jeroyang/ncgocr.
Results: Named concept gene ontology concept recognizer (NCGOCR) achieved 0.804 precision and 0.715 recall by correct recognition of the non-standard mentions of the GO concepts.
Discussion: The lack of consensus on GO naming causes diversity in the GO mentions in biomedical manuscripts. The high performance is owed to the stability of the composing GO concepts and the lack of variance in the spelling of named concepts.
Conclusion: NCGOCR reduced the arduous work of GO annotation and amended the process of searching for the biomarkers or drug targets, leading to improved biomarker development and greater success in precision medicine.
Background
Introduction
In precision medicine, the individual genomic variability is emphasized in prevention, screening, diagnosis and treatment (1, 2). In this regard, the discovery of new biomarkers or drug targets using genome-wide methods requires the support of bioinformatics tools. The gene ontology (GO), composed of three ontologies—biological process, molecular function and cellular component—standardizes the terminology of the gene functions and is extensively used to analyze the results of high-throughput and microarray experiments (3, 4). The data of GO annotation containing the genes and associated gene functions are manually collected from biomedical publications (3, 5). By examining the literature, a few well-trained curators, having knowledge of both biology and GO terminology, established the relationship between the gene and GO concept (5, 6). Although the results acquired by experts who perform GO annotation can be applied to similar genes in other species through the use of software, manual biocuration obstructs the processing of the exponentially growing number of biomedical literature (5, 7, 8).
In relation to the foregoing, the gene ontology concept recognition (GOCR) is the basal component in automatic GO annotation. Given a short paragraph of a biomedical paper, the intuitive approach of GO annotation is to recognize the presence of a gene and GO concept, and then confirm the relationship between them. In the GO annotation tracks of BioCreative I and BioCreative IV, the teams focused most of their efforts to optimize their GOCR systems. Nevertheless, the results remained unsatisfactory (8, 9).
The issues with GOCR are caused by limited synonyms and the limited training data. Some commonly used synonyms of a concept discovered in the annotation are not included in the official synonyms provided by the Gene Ontology Consortium. For example, the term `diurnal cycle’ is not in the list of synonyms of the concept GO:0007623 `circadian rhythm’ because the definitions of these two terms are not the same; only a `diurnal cycle’ having endogenous nature can be called a `circadian rhythm’. While the GO maintainers avoid using `diurnal cycle’, some of the authors use these two terms interchangeably. Automatically marking the term `diurnal cycle’ as a synonym of GO:0007623 `circadian rhythm’ is not difficult for a machine learning-based GOCR system. However, four GO concepts contain the term `circadian rhythm’. If there is not an efficient way to connect the concepts together, we need four independent training examples to update the synonyms of these four concepts. The basic idea of named concept gene ontology concept recognizer (NCGOCR) is to introduce named concepts to handle the highly variable surface names of GO concepts and keep the core definition of each GO concept simple.
Related works
The concept recognition tools, which are integrated with GO, are listed in Table 1. The system Whatizit, using exact string match considering morphological variations, formed the baseline of GOCR (10). The hybrid annotator, Neji, using both dictionary-matching and machine-learning approaches, outperformed Whatizit on the CRAFT corpus (11). The NCBO Annotator uses dictionary-based annotation and expands the annotations using several rules (12). The widely known MetaMap from the National Library of Medicine is best designed for extracting the concepts in the UMLS Metathesaurus (13, 14). The ConceptMapper from the IBM Watson Research Center is equivalent to MetaMap and is more configurable (15).
| System name . | Description . | Ontologies . |
|---|---|---|
| Whatizit (10) | • Web service • Find the exact matching of GO concept and considering morphological variability | Chemical, disease, UMLS, drugs, GO |
| Neji (11) | • Using both dictionary-matching and machine-learning approaches. • Using Gimli, based on conditional random fields model | General purpose |
| NCBO Annotator (12) | • Dictionary-based annotation • Expands the annotations by using: ◆ transitive properties of the ontology ◆ semantic similarity between the concepts ◆ known mappings between different ontology sources. | UMLS, NCBO BioPortal, GO and others |
| MetaMap (National Library of Medicine) (13, 14) | • Tolerate complex match and partial match of noun phrases • Parse the text into noun phrases then: ◆ generate the variants of the noun phrase ◆ obtain the candidate set from Metathesaurus ◆ select the best mapping according to the evaluation function | UMLS, GO and others |
| ConceptMapper (IBM Watson Research Center) (15) | • Highly configurable token-based dictionary lookup • Equivalent to MetaMap | General purpose |
| System name . | Description . | Ontologies . |
|---|---|---|
| Whatizit (10) | • Web service • Find the exact matching of GO concept and considering morphological variability | Chemical, disease, UMLS, drugs, GO |
| Neji (11) | • Using both dictionary-matching and machine-learning approaches. • Using Gimli, based on conditional random fields model | General purpose |
| NCBO Annotator (12) | • Dictionary-based annotation • Expands the annotations by using: ◆ transitive properties of the ontology ◆ semantic similarity between the concepts ◆ known mappings between different ontology sources. | UMLS, NCBO BioPortal, GO and others |
| MetaMap (National Library of Medicine) (13, 14) | • Tolerate complex match and partial match of noun phrases • Parse the text into noun phrases then: ◆ generate the variants of the noun phrase ◆ obtain the candidate set from Metathesaurus ◆ select the best mapping according to the evaluation function | UMLS, GO and others |
| ConceptMapper (IBM Watson Research Center) (15) | • Highly configurable token-based dictionary lookup • Equivalent to MetaMap | General purpose |
| System name . | Description . | Ontologies . |
|---|---|---|
| Whatizit (10) | • Web service • Find the exact matching of GO concept and considering morphological variability | Chemical, disease, UMLS, drugs, GO |
| Neji (11) | • Using both dictionary-matching and machine-learning approaches. • Using Gimli, based on conditional random fields model | General purpose |
| NCBO Annotator (12) | • Dictionary-based annotation • Expands the annotations by using: ◆ transitive properties of the ontology ◆ semantic similarity between the concepts ◆ known mappings between different ontology sources. | UMLS, NCBO BioPortal, GO and others |
| MetaMap (National Library of Medicine) (13, 14) | • Tolerate complex match and partial match of noun phrases • Parse the text into noun phrases then: ◆ generate the variants of the noun phrase ◆ obtain the candidate set from Metathesaurus ◆ select the best mapping according to the evaluation function | UMLS, GO and others |
| ConceptMapper (IBM Watson Research Center) (15) | • Highly configurable token-based dictionary lookup • Equivalent to MetaMap | General purpose |
| System name . | Description . | Ontologies . |
|---|---|---|
| Whatizit (10) | • Web service • Find the exact matching of GO concept and considering morphological variability | Chemical, disease, UMLS, drugs, GO |
| Neji (11) | • Using both dictionary-matching and machine-learning approaches. • Using Gimli, based on conditional random fields model | General purpose |
| NCBO Annotator (12) | • Dictionary-based annotation • Expands the annotations by using: ◆ transitive properties of the ontology ◆ semantic similarity between the concepts ◆ known mappings between different ontology sources. | UMLS, NCBO BioPortal, GO and others |
| MetaMap (National Library of Medicine) (13, 14) | • Tolerate complex match and partial match of noun phrases • Parse the text into noun phrases then: ◆ generate the variants of the noun phrase ◆ obtain the candidate set from Metathesaurus ◆ select the best mapping according to the evaluation function | UMLS, GO and others |
| ConceptMapper (IBM Watson Research Center) (15) | • Highly configurable token-based dictionary lookup • Equivalent to MetaMap | General purpose |
Funk et al. (16) explored the NCBO Annotator, MetaMap and ConceptMapper and systemically adjusted their parameters for the best results in the GOCR. Groza et al. (17) advanced the use of ConceptMapper in the GOCR by integrating the methods based on case-sensitivity matching and term information gain. Funk et al. (18) provided the ConceptMapper with a rule-based method to generate the synonyms of the GO concepts. Consequently, an evident improvement was gained as indicated by an F-measure of 0.636 (0.640 precision, 0.632 recall) on the CRAFT corpus.
Other text-mining systems facilitate GO annotation without identifying the exact locations of the GO concepts. The famous Textpresso, developed by WormBase, is a search engine utilizing keywords and ontology-like hierarchical categories. It helps the curators to narrow down the valuable documents efficiently (19, 20). The powerful GOCat, the leading system in BioCreative IV, takes advantage of previously annotated sentences in the knowledge database. It then assigns a list of prevalent GO concepts and their similarity scores by k-nearest neighbors to the input sentence (21). These systems have resolutions at document and sentence level and cannot be evaluated at character level with the same settings in this study.
Previous systems have had difficulty learning synonyms from the training data. Even when the system learned a new synonym of a concept, it cannot comprehend by analogy how to use this synonym in the related concepts. As a result of this deficiency, the NCGOCR was developed in this study to harvest the synonyms of the named concepts in the limited training data and apply the synonyms to multiple concepts. With the boosting step, we maximized the usage of synonyms via the connection made by the named concepts, which is beyond the capabilities of the previous systems. The high precision (0.804) of NCGOCR could markedly reduce the false positive rate in the search for valuable genes and reduce the workload of human curators.
Materials and methods
We use McDonald’s as a metaphor to explain our design. Imagine that there are a lot of products on the menu of McDonald’s. Some products, such as Big Mac, are made with ingredients such as bun, beef patty, shredded lettuce, sauce, cheese and pickle slices. Besides, some products, e.g. Chicken McNuggets, have only one ingredient. The consumers can customize their order by modifying some of the ingredients in the products. If we check the food on each customer’s tray, would it be possible to trace back the original ordered product? GOCR recognizes the GO concepts from the given documents just like we recognize the customers’ orders.
NCGOCR simplifies the representations of the GO concepts by using smaller fragments, called named concepts. NCGOCR represents the input sentences with these named concepts; then, the GO concepts that share named concepts with the candidates are explored. Finally, NCGOCR confirms the existence and location of the concept using a machine-learning algorithm.
Ogren et al. (22) explained the compositional structure of GO quite well. Most of the GO terms heavily share same tokens with other terms or just contain another GO term. This causes the redundancy in the terminology data and makes it difficult to distinguish between concepts in natural language. For example, there are several associated concepts in the GO, such as `gene silencing`, `regulation of gene silencing’, `negative regulation of gene silencing’ and `negative regulation of gene silencing via microRNA’. The two-word term `gene silencing’ is a substring in all of these terms and is itself a GO concept.
The named concept, a concept has a name that is well accepted by scientists, was introduced to simplify the representation of GO concepts and divide the GOCR into two operable tasks (Figure 1). Accordingly, any GO concept could be split into one or more named concepts. For example, the `GO:0016458 gene silencing’ has only one named concept—`gene silencing’. On the other hand, the `GO:0031047 RNA-mediated gene silencing’ has two named concepts—`RNA’ and `gene silencing’. A named concept may be shared in multiple GO concepts similar to how one ingredient may be used in multiple products at McDonald’s.
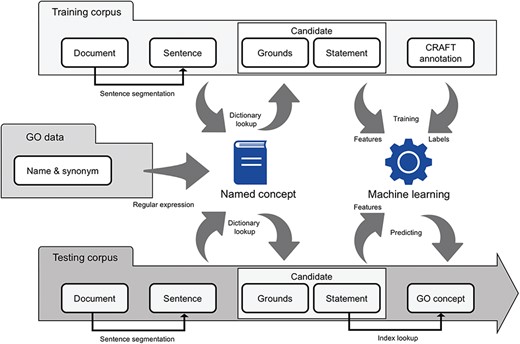
NCGOCR combined dictionary-matching and machine-learning algorithms with named concept.
Problem description
Let Θ = {θ1, θ2… θ|Θ|} be the collection of GO concepts and θ a GO concept. Given the document Γ, the problem is to identify the references to the GO concept, θ, including their start and end positions in the document.
Corpus and resources
The CRAFT corpus 1.0 contains 67 full-text articles associated with manual annotations to multiple biomedical ontologies and terminologies, including GO (23). It provides the location of a reference to a GO concept and the unique identifier (GOID) in the given article.
To make the results of GOCR pertinent, the GO data provided with the CRAFT corpus (23)—containing 24 337 concepts (biological process: 14 361, molecular function: 7980 and cellular component: 2047)—were used in this study.
Generating named concepts and statements from GO data
Apart from the ingredients already listed in the menu of McDonald’s, we have to find out the ingredients—we call them named concepts—of these items by ourselves. With a group of regular expressions (listed in Table S1 in the supplementary data), the name or synonym of a GO concept is decomposed into a product of named concepts and statements (Figure 2). A statement is a concise representation of a GO concept that consists of named concepts. Each name or synonym of a GO concept is represented with statement, s = {m1, m2, … m|s|}, where m represents the named concept, and a unique identifier for each statement (statement ID) is assigned. Using the metaphor of McDonald’s, the GO statements are similar to the products on the menu, which are the standards. A GO concept is a collection of one or multiple statements, just as a cheeseburger may be a Cheeseburger, Double Cheeseburger or Triple Cheeseburger.
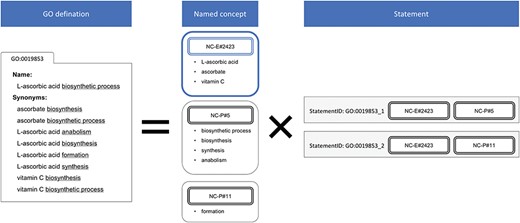
Decomposition of the GO definition. NC-P#5 and NC-P#11 were defined heuristically in our regular expression, and the naïve NC-E#2423 was generated in this process. The redundant nine definitions of GO:0019853 are simplified into two statements. The variant spellings of vitamin C are handled by NC-E#2423.
The regular expressions in Table S1 were designed based on several common fragments in the names or synonyms, usually representative of some actions, such as `regulation’, `positive regulation’, `activity’ and `transport’. These named concepts are called `the pattern’ (NC-P). The surface names of the NC-P are used to cut a name or a synonym and split it into named concepts. The named concepts made by the previous decomposing process, account for the majority of named concepts, are called `the entity’ (NC-E). `The constraint’ (NC-C), representing the constraints of the GO concepts and only presented in some concepts deep down in the hierarchy of GO, were filtered by the NC-C regular expressions beforehand (Table S1). All the surface names of the named concepts were collected as the targets in the dictionary-matching step. For example, GO:0007623 `circadian rhythm’ has only one named concept: `circadian rhythm’ (NC-E); GO:0042752 `regulation of circadian rhythm’ contains `regulation’ (NC-P) and `circadian rhythm’ (NC-E); GO:0042753 `positive regulation of circadian rhythm’ contains `positive regulation’ (NC-P) and `circadian rhythm’ (NC-E); GO:0042754 `negative regulation of circadian rhythm’ contains `negative regulation’ (NC-P) and `circadian rhythm’ (NC-E). These four concepts having the common `circadian rhythm’ (NC-E) will later benefit from the boosting step.
Text processing and candidate generation
A majority (∼97%) of the GO concepts in the CRAFT corpus do not cross the boundaries of sentences, so we made sentence segmentation of the documents as a preprocessing procedure. For a document Γ = {γ1, γ2, …, γ|Γ|}, where γ represents the sentence, the sentence’s start and end positions were tracked. This was performed as well in the succeeding candidate generation steps for the final reporting of GOCR.
The use of word tokenization to match the surface names of the named concepts was abandoned because a named concept usually crosses the boundaries of the tokens. Instead, the Aho–Corasick algorithm, a finite state machine based on a prefix tree, was employed to perform the dictionary-matching task (24). For each input sentence, γ, all the surface names of the known named concepts were located and then gathered in a list of associated named concepts, called `grounds’, Gγ (Figure 3).
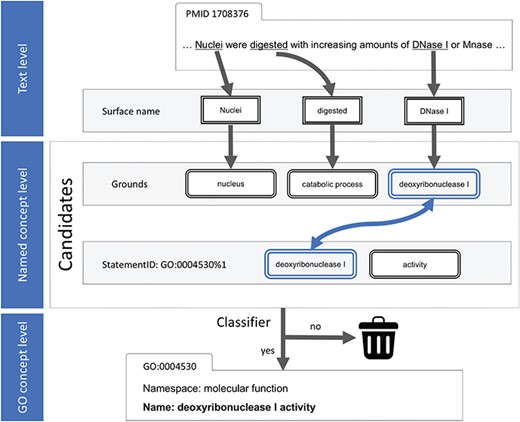
Text processing and machine-learning diagram. On the named concept level, the candidate was generated by the greedy search for at least one common named concept in the grounds and statement.
Thereafter, a candidate—containing a related statement, s, pointing to the associated GO concept—is derived from Gγ. Both s and Gγ are lists of named concepts, where s is from the name or synonyms and Gγ is from the sentence γ. All related statements are thoroughly explored to ascertain if s and Gγ had at least one shared named concept (Figure 3). A candidate is a guess of the GO statement from given named concepts found in the input sentence. Imagine that we checked the customer’s tray and found a fish filet patty, a regular bun and an American cheese (the standard ingredients of Filet-O-Fish). All products on the menu that have any one of the ingredients above will be included as a candidate; the number of the candidates is thus large because of this greedy process.
A boosting step was also applied to enrich both the surface names and statements from the training corpus. If the annotated GO concept in the training corpus has a statement that contains only one named concept, its mention can be intuitively declared as a new surface name linked to this named concept. Otherwise, if the mention of the annotated GO concept contains only one word, a new statement associated with the target GO concept, containing two named concepts, is created. The first named concept is linked to the mention of this single word as its surface name and the second `null’ named concept is linked to nothing. By boosting the statements, a link from the text to GO concept by a newly defined named concept is created, allowing the machine-learning algorithm to hone its effect. For example, the term `diurnal cycle’ matches GO:0007623 `circadian rhythm’ in the training data, the boosting step links the surface name `diurnal cycle’ with the `circadian rhythm’ (NC-E) automatically. Our system can, therefore, utilize this surface name for all the four concepts containing `circadian rhythm’ (NC-E). In the metaphor of McDonald’s, a boosting step is to learn to recognize the variant shapes of the ingredients. If we learned that the melted cheese is actually the same cheese in the example of Filet-O-Fish, this knowledge would be applied to all the products with cheese on the menu. We could recognize a cheeseburger with melted cheese without seeing an example in the training data.
Machine-learning process
Six features in three categories for each candidate were designed in the machine-learning process. The features in the concept category contained the statement ID and the namespace of this concept. The features in the evidence category are related to the lowercase mention that was found in the given sentence, including its span, prefix and suffix. Moreover, saturation, |$\theta =\frac{\left|G\,\cap\, s\right|}{\left|s\right|}$|, is the ratio of the number of matched named concepts to the count of the named concepts in the target statement. For example, in the sentence `Nuclei were digested with increasing amounts of DNase I or MNase.’, obtained from PubMed unique identifier (PMID):17083276, three surface names are found: nuclei, digested and deoxyribonuclease I (DNase I), which linked to three named concepts—nucleus, catabolic process and DNase I, respectively. Accordingly, these named concepts are denoted as being in the grounds G = {nucleus, catabolic process, deoxyribonuclease I}. In Table 2, the example candidate is associated with the statement GO:0004530_1, s = {deoxyribonuclease I, activity}. There is only one named concept {deoxyribonuclease I} in s presented in G. Hence, the saturation is 0.5 (1 in 2).
| Feature . | Example . | Category . |
|---|---|---|
| Statement ID | GO:0004530_1 {deoxyribonuclease I, activity} | Concept |
| Namespace | molecular function | |
| Length | 7 (of the mention `DNase I’) | Evidence |
| First three characters | DNA (of the mention `DNase I’) | |
| Last three characters | e_i (of the mention `DNase I’) | |
| Saturation | 0.5 | Bias |
| Feature . | Example . | Category . |
|---|---|---|
| Statement ID | GO:0004530_1 {deoxyribonuclease I, activity} | Concept |
| Namespace | molecular function | |
| Length | 7 (of the mention `DNase I’) | Evidence |
| First three characters | DNA (of the mention `DNase I’) | |
| Last three characters | e_i (of the mention `DNase I’) | |
| Saturation | 0.5 | Bias |
This example candidate and its features were generated from the sentence `Nuclei were digested with increasing amounts of DNase I or MNase.’
| Feature . | Example . | Category . |
|---|---|---|
| Statement ID | GO:0004530_1 {deoxyribonuclease I, activity} | Concept |
| Namespace | molecular function | |
| Length | 7 (of the mention `DNase I’) | Evidence |
| First three characters | DNA (of the mention `DNase I’) | |
| Last three characters | e_i (of the mention `DNase I’) | |
| Saturation | 0.5 | Bias |
| Feature . | Example . | Category . |
|---|---|---|
| Statement ID | GO:0004530_1 {deoxyribonuclease I, activity} | Concept |
| Namespace | molecular function | |
| Length | 7 (of the mention `DNase I’) | Evidence |
| First three characters | DNA (of the mention `DNase I’) | |
| Last three characters | e_i (of the mention `DNase I’) | |
| Saturation | 0.5 | Bias |
This example candidate and its features were generated from the sentence `Nuclei were digested with increasing amounts of DNase I or MNase.’
The benchmark from the CRAFT corpus contains a five-tuple for the results of the GOCR: the PMID, GOID, start position, end position and annotated text, which are not directly suitable for the machine-learning process. Accordingly, a candidate is marked positive if the two following conditions were satisfied: (i) the GOID of the candidate matches the one found in the benchmark and (ii) the text span of the candidate overlaps that in the benchmark with the exact GOID. Although some character shift was allowed in the training step to lower the criteria for positive results, a stringent setting on evaluation was retained. In this regard, the random forest classifier was employed along with the default setting of the parameters from the scikit-learn module (version 0.18.1) of Python (25).
Evaluation
To evaluate the proposed system, we employed 10-fold cross-validation. The CRAFT corpus was randomly divided into ten groups at the document level—where nine of these groups were united as the training dataset—and the remaining group was handled as the testing dataset for each epoch of cross-validation (Table 3). Thus, the testing dataset was neither used in the boosting nor machine-learning step.
Consequently, the proposed system was carefully and thoroughly validated with the benchmark—i.e. a report of the annotation was counted as true positive only if the start position, end position and GOID all matched those of the benchmark. Finally, the micro-averages of the recall, precision and F-measure of the system were calculated.
Results
Evaluation of the NCGOCR
In the evaluation of the NCGOCR system, the following micro-averages were obtained: precision, 0.804; recall, 0.715 and F-measure, 0.757. As shown in Figure 4, the NCGOCR achieved evident improvements in both precisions and recalls of all namespaces, especially in the biological process and molecular function. All the specific values of the results of the NCGOCR evaluation are listed in Table S2 in the supplementary data. The frequency distribution of the named concepts generated from these GO concepts is shown in Figure S1.
| Number of . | Average . | Standard deviation . |
|---|---|---|
| Documents | 6.7 | 0.5 |
| Sentences | 2164.6 | 329.0 |
| GO concepts | 2944.7 (100%) | 637.0 |
| Biological process | 1691.3 (57.4%) | 412.9 |
| Molecular function | 418.0 (14.2%) | 198.6 |
| Cellular component | 835.4 (28.4%) | 320.7 |
| Unique GO concepts | 284.3 (100%) | 37.1 |
| Biological process | 174.5 (61.1%) | 25.8 |
| Molecular function | 57.8 (20.2%) | 8.8 |
| Cellular component | 53.5 (18.7%) | 13.4 |
| Number of . | Average . | Standard deviation . |
|---|---|---|
| Documents | 6.7 | 0.5 |
| Sentences | 2164.6 | 329.0 |
| GO concepts | 2944.7 (100%) | 637.0 |
| Biological process | 1691.3 (57.4%) | 412.9 |
| Molecular function | 418.0 (14.2%) | 198.6 |
| Cellular component | 835.4 (28.4%) | 320.7 |
| Unique GO concepts | 284.3 (100%) | 37.1 |
| Biological process | 174.5 (61.1%) | 25.8 |
| Molecular function | 57.8 (20.2%) | 8.8 |
| Cellular component | 53.5 (18.7%) | 13.4 |
| Number of . | Average . | Standard deviation . |
|---|---|---|
| Documents | 6.7 | 0.5 |
| Sentences | 2164.6 | 329.0 |
| GO concepts | 2944.7 (100%) | 637.0 |
| Biological process | 1691.3 (57.4%) | 412.9 |
| Molecular function | 418.0 (14.2%) | 198.6 |
| Cellular component | 835.4 (28.4%) | 320.7 |
| Unique GO concepts | 284.3 (100%) | 37.1 |
| Biological process | 174.5 (61.1%) | 25.8 |
| Molecular function | 57.8 (20.2%) | 8.8 |
| Cellular component | 53.5 (18.7%) | 13.4 |
| Number of . | Average . | Standard deviation . |
|---|---|---|
| Documents | 6.7 | 0.5 |
| Sentences | 2164.6 | 329.0 |
| GO concepts | 2944.7 (100%) | 637.0 |
| Biological process | 1691.3 (57.4%) | 412.9 |
| Molecular function | 418.0 (14.2%) | 198.6 |
| Cellular component | 835.4 (28.4%) | 320.7 |
| Unique GO concepts | 284.3 (100%) | 37.1 |
| Biological process | 174.5 (61.1%) | 25.8 |
| Molecular function | 57.8 (20.2%) | 8.8 |
| Cellular component | 53.5 (18.7%) | 13.4 |
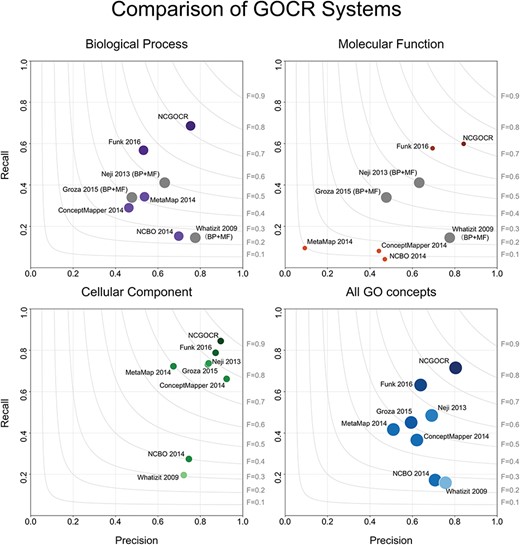
Comparison of the precision and recall of the GOCR systems. The surface area of the circles represents the amount of the annotations in the benchmark. BP: biological process and MF: molecular function.
Analysis of the system components
Each component was eliminated from the system to evaluate its contribution individually (Table 4). One of the key components is boosting, which performed an essential task in increasing the recall. Although the boosting process required a small loss in precision (from 0.842–0.804), it doubled the recall (from 0.344–0.715) and greatly improved the F-measure (from 0.488–0.757). Moreover, among the three feature categories, the evidence feature is found to be more important than the other categories. Nevertheless, the single most important feature is the statement ID, which contributed 0.060 to the system F-measure.
| Removed component . | Precision . | Recall . | F-measure . | Delta to the full system . |
|---|---|---|---|---|
| Full system (baseline) | 0.804 | 0.715 | 0.757 | 0.000 |
| Boosting | 0.842 | 0.344 | 0.488 | −0.269 |
| Concept features | 0.763 | 0.657 | 0.706 | −0.051 |
| Statement ID | 0.734 | 0.665 | 0.697 | −0.060 |
| Namespace | 0.798 | 0.718 | 0.755 | −0.002 |
| Evidence features | 0.690 | 0.657 | 0.672 | −0.085 |
| Length | 0.787 | 0.698 | 0.739 | −0.018 |
| Text | 0.793 | 0.714 | 0.751 | −0.006 |
| Prefix and suffix* | 0.782 | 0.712 | 0.745 | −0.012 |
| Saturation feature | 0.785 | 0.696 | 0.738 | −0.019 |
| Removed component . | Precision . | Recall . | F-measure . | Delta to the full system . |
|---|---|---|---|---|
| Full system (baseline) | 0.804 | 0.715 | 0.757 | 0.000 |
| Boosting | 0.842 | 0.344 | 0.488 | −0.269 |
| Concept features | 0.763 | 0.657 | 0.706 | −0.051 |
| Statement ID | 0.734 | 0.665 | 0.697 | −0.060 |
| Namespace | 0.798 | 0.718 | 0.755 | −0.002 |
| Evidence features | 0.690 | 0.657 | 0.672 | −0.085 |
| Length | 0.787 | 0.698 | 0.739 | −0.018 |
| Text | 0.793 | 0.714 | 0.751 | −0.006 |
| Prefix and suffix* | 0.782 | 0.712 | 0.745 | −0.012 |
| Saturation feature | 0.785 | 0.696 | 0.738 | −0.019 |
*Remove two features: the first three characters and last three characters of the mention.
| Removed component . | Precision . | Recall . | F-measure . | Delta to the full system . |
|---|---|---|---|---|
| Full system (baseline) | 0.804 | 0.715 | 0.757 | 0.000 |
| Boosting | 0.842 | 0.344 | 0.488 | −0.269 |
| Concept features | 0.763 | 0.657 | 0.706 | −0.051 |
| Statement ID | 0.734 | 0.665 | 0.697 | −0.060 |
| Namespace | 0.798 | 0.718 | 0.755 | −0.002 |
| Evidence features | 0.690 | 0.657 | 0.672 | −0.085 |
| Length | 0.787 | 0.698 | 0.739 | −0.018 |
| Text | 0.793 | 0.714 | 0.751 | −0.006 |
| Prefix and suffix* | 0.782 | 0.712 | 0.745 | −0.012 |
| Saturation feature | 0.785 | 0.696 | 0.738 | −0.019 |
| Removed component . | Precision . | Recall . | F-measure . | Delta to the full system . |
|---|---|---|---|---|
| Full system (baseline) | 0.804 | 0.715 | 0.757 | 0.000 |
| Boosting | 0.842 | 0.344 | 0.488 | −0.269 |
| Concept features | 0.763 | 0.657 | 0.706 | −0.051 |
| Statement ID | 0.734 | 0.665 | 0.697 | −0.060 |
| Namespace | 0.798 | 0.718 | 0.755 | −0.002 |
| Evidence features | 0.690 | 0.657 | 0.672 | −0.085 |
| Length | 0.787 | 0.698 | 0.739 | −0.018 |
| Text | 0.793 | 0.714 | 0.751 | −0.006 |
| Prefix and suffix* | 0.782 | 0.712 | 0.745 | −0.012 |
| Saturation feature | 0.785 | 0.696 | 0.738 | −0.019 |
*Remove two features: the first three characters and last three characters of the mention.
Discussion
The lack of consensus on naming, particularly about the terminologies used for GO concepts, is making GOCR difficult. The diversity in the mentions of a GO concept in the context reflects the uncertainty on naming, which is a consequence of the compositional structure of GO. Evidently, the names of the GO concepts and their compositional structures are too complex to fit in the human brain. Thus, whenever a manuscript being written describes a complex GO concept, such as `negative regulation of gene silencing via microRNA’, its author might coin another description and use it in the manuscript instead of the multi-word term. In the preceding example, it is evident that although scientists are aware of the same GO concept, a consensus on its name has not been reached. Consequently, this scenario creates varying presentations of GO concepts in documents and thrusts GOCR towards a difficult task. In contrast, named entity recognition with normalization—an easier form of ontology recognition—locates named entities inside a given text and maps these into a predefined list of interests. In named entity recognition, the entities, such as names, locations or companies are treated as flat structures. Thus, each named entity is not related to the other entities and has a consensual name. Among the three namespaces of GO, the recognition of the cellular component is most similar to named entity recognition. The concepts in the cellular component are proper nouns with a consensual name, indicating different levels of the cell structures, such as `spindle microtubule’, `microtubule’ and `cytoskeleton component’. In Figure 4 and Table S2, the cellular component has the highest values in precision, recall and F-measure in all systems because less ambiguity in naming makes GOCR in the cellular component easier than that in the other two namespaces.
A widely known problem in GOCR is the lack of training data and only ∼5% of the GO concepts are utilized in the CRAFT corpus. On this account, how does the NCGOCR, based on only 5% of the GO concepts, work on the remaining 95% of the concepts?
Firstly, in the dictionary-matching step, the training data multiply their effects via the shared named concepts among the GO concepts. The new surface names gathered from the training corpus in the boosting step—mostly plural forms, abbreviations or alternative spellings—are spread to many GO concepts with the same named concepts. Accordingly, boosting the surface name and statement performed an important function in increasing the recall. In fact, the 5% of the GO concepts in the CRAFT corpus covers ∼14% of the named concepts, and this 14% of named concepts are widely used in 74% of all GO concepts. Secondly, all training data were concentrated on building one binary classifier, which determined whether the candidate had a valid GO concept or not. The lower bound performance on never-seen GO concepts could be recognized from the list in Table 4. The system—blind to the processing of the GO concept—had an F-measure of 0.706 (−0.051). Based on both dictionary-matching and machine-learning steps, the NCGOCR is stable and reliable during the changing of the corpora. In fact, the majority of the GO concepts are barely used (or in some cases never used) in the annotation. Since the annotations per concept follow the Zipfian law (22), the size of the training corpus slowly increases the coverage of the GO concepts. A corpus with high coverage of GO concepts may be impossible to obtain.
Another major problem deals with the migration of GO (26). There are more than 40 000 GO concepts today, and the number is continuously growing. How does NCGOCR, which is based on the 2012 corpus, work on the present projects? In the dictionary-matching step, the relationship between the surface names and named concepts is stable over time. Thus, the named concept, a concept having a consensual name by definition, should not frequently change during the evolution of the GO because the name is embedded within the language and society of the scientists who use it. In the machine-learning step, the logic to form a concept from the features, which was learned by the classifier, should also be preserved over time. Whenever there is no newer training material other than the CRAFT corpus, it is presumed that the NCGOCR can operate well with different versions of the GO data by modifying the named concepts and statements without the need of retraining the classifier.
Conclusion
Variance in spelling is the fundamental difficulty when mapping natural language mentions into a controlled vocabulary. The concept recognition work on GO is more difficult than those on the gene, chemical and disease names because of the compositional nature of GO. The gradation of text, named concept and GO concept elucidates the two layers of these kind of problems: the variation in spelling and the existence of a concept. The NCGOCR accurately solved the problems in these two layers through the dictionary-matching and machine-learning algorithms. The improvements in the overall F-measure and precision, especially on the biological process and molecular function, sufficiently provided the information needed in the field of precision medicine.
Funding
Ministry of Science and Technology, Taiwan (MOST 103-2221-E-006-254-MY2).
Conflict of interest. None declared.
Database URL:https://github.com/jeroyang/ncgocr
References



