-
PDF
- Split View
-
Views
-
Cite
Cite
Vincent Henry, Ivan Moszer, Olivier Dameron, Laura Vila Xicota, Bruno Dubois, Marie-Claude Potier, Martin Hofmann-Apitius, Olivier Colliot, INSIGHT-preAD Study Group, Converting disease maps into heavyweight ontologies: general methodology and application to Alzheimer’s disease, Database, Volume 2021, 2021, baab004, https://doi.org/10.1093/database/baab004
Close - Share Icon Share
Abstract
Omics technologies offer great promises for improving our understanding of diseases. The integration and interpretation of such data pose major challenges, calling for adequate knowledge models. Disease maps provide curated knowledge about disorders’ pathophysiology at the molecular level adapted to omics measurements. However, the expressiveness of disease maps could be increased to help in avoiding ambiguities and misinterpretations and to reinforce their interoperability with other knowledge resources. Ontology is an adequate framework to overcome this limitation, through their axiomatic definitions and logical reasoning properties. We introduce the Disease Map Ontology (DMO), an ontological upper model based on systems biology terms. We then propose to apply DMO to Alzheimer’s disease (AD). Specifically, we use it to drive the conversion of AlzPathway, a disease map devoted to AD, into a formal ontology: Alzheimer DMO. We demonstrate that it allows one to deal with issues related to redundancy, naming, consistency, process classification and pathway relationships. Furthermore, we show that it can store and manage multi-omics data. Finally, we expand the model using elements from other resources, such as clinical features contained in the AD Ontology, resulting in an enriched model called ADMO-plus. The current versions of DMO, ADMO and ADMO-plus are freely available at http://bioportal.bioontology.org/ontologies/ADMO.
Introduction
Systems medicine disease maps (DMs; see Appendice 1 for abbrevations) provide curated and integrated knowledge on pathophysiology of disorders at the molecular and phenotypic levels (1). Based on a systems biology approach, they describe all biological physical entities (i.e. gene, mRNA, protein and metabolite) in their different states (e.g. phosphorylated protein, molecular complex and degraded molecule) and the interactions between them. Their relations are represented as molecular interactions (as well as covalent modifications) organized in pathways, which encode the transitions between participants’ states as processes (1, 2). Most advanced DM projects focus on Parkinson’s disease (3), cancer (4), rheumatoid arthritis (5, 6), asthma (7), atherosclerosis (8), macrophage activation transduction signaling (9) and Alzheimer’s disease (AD) (10).
AD is a progressive neurodegenerative disorder of the brain, which was first described in 1906. The intense activity of AD research constantly generates new data and knowledge on AD-specific molecular and cellular processes (a Medline search for ‘Alzheimer’s disease’ results in over 115 000 articles, as of December 2019). However, the complexity of AD pathophysiology is still imperfectly understood (11). These 110 years of efforts have essentially resulted in one dominant paradigm to underline the causes of AD: the amyloid cascade (12). Nevertheless, therapeutics targeting this pathway failed to lead to curative outcome for humans, leading to the need for additional hypotheses (13). Briefly, several approaches have been pursued in order to target the amyloid metabolic cascade for the treatment of AD (14). Among them, there have been treatments targeting BACE-1 that proved to lower Aβ production and brain amyloid load in animal models (15) but did not show any improvement in cognition in clinical trials (16) (lanabecestat), or even worsened symptoms (17). Similar results were found for drugs that targeted the γ-secretase, for immunization approaches, or for treatment with monoclonal antibodies. This could be explained by the fact that Aβ accumulation is a gradual process that takes many years to occur and is linked to changes in the macro- and microenvironment of the brain and the neurons—including neuroinflammation, alterations in endolysosomal trafficking and tau accumulation—membrane cholesterol changes. Therefore, stopping the amyloid cascade when the environment is already altered might not be sufficient to improve cognitive deficits or to stop the cognitive decline. In conclusion, treatments should potentially start before the apparition of cognitive signs and should likely be combined with treatments targeting other mechanisms.
Since the turn of the century, omics technologies (e.g. genomics, transcriptomics, proteomics, phosphorylomics and metabolomics) lead to a more comprehensive characterization of biological systems and diseases. The production of omics data in AD research thereby opens promising perspectives to identify alternatives to the amyloid cascade paradigm. There is a clear need to integrate the amyloid cascade as a component of the whole organ-wide dysregulation occurring in AD, rather than treating it as an isolated component. Therefore, a model should be built that integrates tau, neuroinflammation, cholesterol metabolism, insulin resistance, neuronal degeneration and all the other known pathways involved in AD. The current challenge is to connect and integrate these data in an appropriate way.
AlzPathway is a DM developed for AD (10). Although very rich in AD-specific pathophysiology information (it describes 1347 biological physical entities, 129 phenotypes, 1070 biochemical reactions and 26 pathways), this resource does not provide sufficient formalism to adequately interlink current knowledge and omics data: it would thus benefit from a refined level of description, able to cope with the complex modeling of disease systems and the diversity of measurements from biomedical experiments. This lack of formalism is inherent to all DMs.
The information contained in DM is stored in syntactic formats developed for systems biology: the Systems Biology Graphical Notation (18), the modified Edinburg Pathway Notation (mEPN) (19) and the Systems Biology Markup Language (SBML) (20). While syntactic formats are able to index information and can be managed by different applications such as MINERVA (21) or NaviCell (22), they are not expressive enough to define explicit relationships and formal descriptions, leading to possible errors and misinterpretations (e.g. reaction ‘re1178’ is describing the translation of the IL1B gene into IL1B mRNA; this description does not allow to interpret whether re1178 is a transcription instead of a translation or whether IL1B gene is a transcript instead of a gene). For AlzPathway, this defect in expressiveness results in a lack of (i) hierarchy and disjunction between species (e.g. between ‘Protein’ and ‘phosphorylated Protein’ or between ‘Protein’ and ‘RNA’, respectively), (ii) formal definition of entities (such as phenotypes), (iii) formal relationships between reactions and pathways, (iv) uniformity of entities’ naming (e.g. complexes that are labelled by their molecular components or by a common name) and (v) consistency between reactions and their participants (e.g. translation of genes instead of transcripts).
Compared to syntactic formats, the Web Ontology Language (OWL), a semantic format used in ontologies, has higher expressiveness (23) and was designed to support knowledge and data integration. Moreover, OWL combines high expressivity and logical constraints to ensure the consistency of the resource (24). It is thus a good candidate to overcome the previous limitations. An ontology is an explicit specification of a set of concepts and their relationships, represented in a knowledge graph in semantic format. Ontologies provide a formal naming and definition of the types (i.e. the classes; see Appendice 2 for glossary) and interrelationships between entities (i.e. the properties) that exist for a particular domain. Moreover, knowledge and data managed by an ontology benefit from their logical semantics and axiomatic properties (e.g. class disjunction, cardinality, existentiality, universality), which supports automatic control of consistency and additional information inferences (including hierarchy and relationships) (25).
In the biomolecular domain, the Gene Ontology (GO) provides the community with the largest set of controlled vocabulary to index and share data (26). WikiPathways (27) and the Systems Biology Ontology (SBO (28)) also provide controlled vocabulary hierarchies. But none of these ontologies provide enough specificity for AD pathophysiology. In the AD domain, the Alzheimer’s Disease Ontology (ADO) (29) organizes information describing clinical, experimental and molecular features in OWL format for text mining. However, the description of the molecular systems of ADO is less specific than that of AlzPathway.
In this paper, we propose the Disease Map Ontology (DMO), an ontological upper model able to drive the conversion of a DM into a formal ontology. We then apply it to convert AlzPathway into an OWL ontology, which we call the Alzheimer Disease Map Ontology (ADMO). Finally, we show that ADMO can be connected with ADO into ADMO-plus, a resource able to store and interconnect biomedical data. These different steps are summarized in Figure 1.
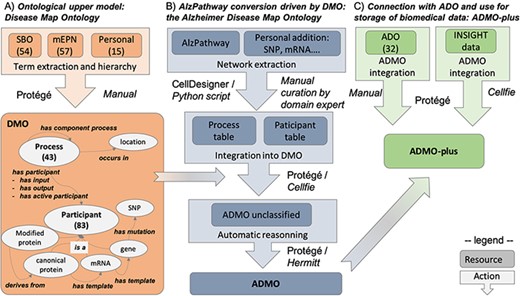
Summary of the workflow for AlzPathway conversion in OWL, from the DMO design to ADO instantiation and data integration. (A) DMO design. (B) AlzPathway export into a structured table and its integration with DMO, resulting in ADMO. (C) Integration of ADO and biomedical experiment data resulting in ADMO-plus. This is not a pipeline, but a step-by-step process, in which manual and automatic steps are specified. Specifically, for each step, we indicate whether it was done manually (Manual) or mention which tool was used to do it automatically (Protégé, Cell designer, Python script, Cellfie, Hermitt).
Ontological upper model: DMO
The first task (Figure 1A) aimed at designing a generic ontological upper model able to drive the conversion of the specific content of a DM (in our case AlzPathway). In an expressive ontology, the relationships are not only links between classes, but also logical constraints (i.e. axioms) that are inherited by all their descendants (subclasses). Thus, the choices of axioms that support high level classes and their properties are key elements for the usefulness of the model.
Design of DMO classes
SBO (28) is a terminology that provides a set of classes commonly used to index information in SBML format. These classes conceptualize biological entities with a suitable balance between genericity and specificity in order to improve the genericity of representation despite the diversity of reactions: thus systemic models can be adequately represented using few classes, while preserving a satisfactory level of accuracy, similarly to the BioModels representation (30). To build the DMO ontological model, we first selected SBO terms from ‘process’ or ‘material entity’ classes that fit with DM content, specifically those corresponding to the reaction types present in the map legend. This resulted in 54 terms: 37 reaction types and 17 molecule types, respectively. Then, we relied on the vocabulary used to define components’ shapes in the graphical format mEPN, to complete the SBO class set with molecular states that fit with DM knowledge (e.g. phosphorylated or truncated). Following class selection from SBO and mEPN, we designed a class hierarchy between them. Classes related to participants were separated into two hierarchies: one describing their biochemical properties such as polypeptide chains, simple chemicals, genes or non-covalent complexes, one describing the state of participants such as native form, phosphorylated or truncated. We systematically added disjointness constraints between the generic sibling subclasses of participants in the biochemical property hierarchy in order to ensure that process participants belong to only one set (e.g. a gene cannot be a protein and reciprocally). We did not apply the same rule to the state hierarchy as, for instance, a truncated protein could also be phosphorylated. Classes related to processes were also hierarchized without disjointness constraints as a reaction may refer to different processes (e.g. a transfer is an addition and a removal).
Design of DMO properties
Properties consistent with a systems approach (i.e. has_part, has_component, has_component_process, has_participant, has_input, has_output, has_active_participant and their respective inverse properties) were selected from the upper-level Relation Ontology (RO) (31). Then, we enriched the formal definition of our set of process classes with these properties and associated cardinalities to link processes and participants with relationships in description logic (e.g. a transcription has at least one gene as input and at least one mRNA as output; a protein complex formation has at least two proteins as input and at least one protein complex as output).
Finally, four other properties were added: occurs_in (a property selected from RO) to link a process to its respective location, derives_from to link a modified protein to its initial form, has_template (sub-property of derives_from) to link a mRNA to its related gene or a protein to its related mRNA, and has_mutation (sub-property of has_part) to link a gene to its possible mutations.
DMO design results
The design of the DMO upper ontological model based on SBO, mEPN, RO and de novo additions resulted in 143 classes (43 processes’ subclasses and 83 participants’ subclasses) and 14 properties formally defined by 188 logical axioms in description logic (Figure 2). This model is based on a simple pattern as our knowledge graph involves only four types of properties (and their inverse properties): (i) the is_a (subclass_of) standard property, (ii) the has_part standard property and its sub-properties has_component, has_component_process and has_mutation (iii) the has_participant property and its sub-properties has_input, has_output and has_active_participant and (iv) the location property occurs_in.
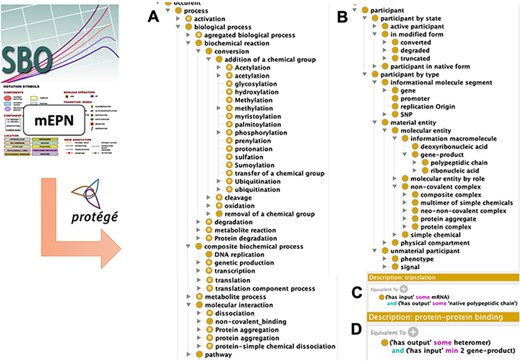
Disease Map Ontology (DMO) model design. Term of classes were extracted from the Systems Biology Ontology (SBO) and the modified Edinburg Pathway Notation format (mEPN) into Protégé. Classes were hierarchized as subclasses of process (A) or participant (B). Using property terms from the Relation Ontology (RO), classes were formally defined in description logic, as illustrated in the case of transcription (C) and protein complex formation (D) processes.
AlzPathway conversion driven by DMO: the ADMO
DMO was designed to integrate DM knowledge as subclasses and manage its consistency and formalism. Here, we demonstrate its use in the case of the conversion of the AlzPathway DM into a formal ontology, ADMO (Figure 1B).
Extraction of AlzPathway contents
AlzPathway information, contained in the original SBML file (32), was exported using CellDesigner (33) in a tabular format, which was further restructured using home-made Python scripts (Supplementary Figure S1). This step involved both manual and automatic transformations. We thus created a table (Supplementary Table S1) in which each biological entity was indexed by one of the DMO participants’ subclasses and all processes were matched with their input, output or active participants. In AlzPathway, reactions have no naming: they are labeled from r1 to r1070. To facilitate human readability, processes were labelled with a concatenation of the type of the reactions and the names of the participants. The table was also supplemented with class annotations corresponding to other information contained in AlzPathway, such as the AlzPathway identifier (ID), and IDs from other knowledge bases like UniProt (34) for participants and KEGG (35) for processes. The table was structured to integrate component information in case of multiplex entities (e.g. protein complexes) or the initial (native) entity in case of modified entities (e.g. phosphorylated or truncated proteins) and location information for processes (e.g. cell type or cell part). The table was then manually curated as described below.
AlzPathway content modification and addition
In AlzPathway, native and modified proteins (e.g. phosphorylated or activated) have the same name and differ only in their graphical shapes. In order to specify these different states, we added a suffix to modified protein labels (e.g. ‘_P’ or ‘_a’ for phosphorylated or activated, respectively).
In AlzPathway, phenotypes are participants. But several of them are named with a process name, pathway label or molecule type (e.g. microglial activation, apoptosis or cytokines, respectively). In order to deal with these ambiguities, 26 phenotypes were reclassified as molecules (e.g. cytokine) or cellular components (e.g. membrane) and 14 names that referred to processes or pathways were changed into processes’ participant names (e.g. ‘apoptosis’ that refers to a process was changed into ‘apoptotic signal’ that refers to a participant). In addition, 5 phenotypes that were named with a pathway name (e.g. apoptosis) were added to the initial set of the 26 AlzPathway’s pathways.
AlzPathway describes a subset of genes, mRNAs and proteins, but not always the whole combination of one given gene, its related mRNAs and proteins. As omics technology can capture data at the genome, transcriptome or proteome levels, we added missing genes, mRNAs or proteins in order to always have the description of the gene, the mRNA and the protein for a same entity described in AlzPathway. Additional entities were linked with the has_template relationships but not linked to reactions when no corresponding knowledge was found in AlzPathway. In such a way, we avoided overinterpretations. This resulted in the addition of 407 genes, 416 mRNAs and 191 proteins.
AlzPathway conversion in OWL format
Then, using the Protégé ontology editor (a free, open-source software that provides a convenient interface to edit OWL files, widely used in research and supported by an international community) (36), the content of the structured table was imported into DMO and converted in OWL using the Protégé Cellfie plugin. AlzPathway’s molecular and phenotypic entities were integrated as subclasses of DMO ‘participant’ classes without redundancies (172 redundant participants were identified). Reactions extracted from AlzPathway were integrated as independent subclasses of the ‘process’ class, without hierarchy. Then, automatic reasoning was used to classify them as subclasses of the DMO upper model process classes depending on their formal definition (see Figure 3a*), independently of their initial types in AlzPathway. The 1065 inferred SubClassOf axioms were added to the ontology.
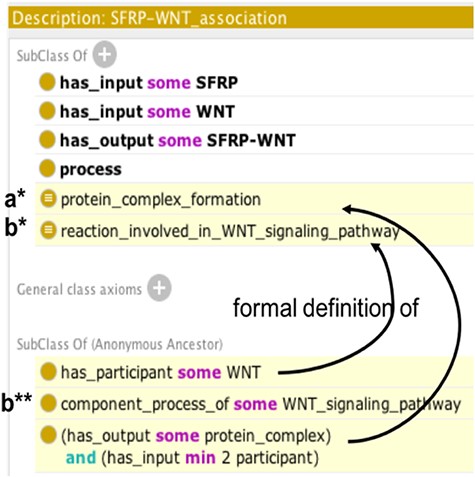
Example of automatic reasoning with Protégé. Asserted axioms are shown in uncolored lines and inferred axioms are highlighted in yellow. Following automatic reasoning, SFRP-WNT heterodimer association is classified as subclass of the ‘protein complex formation’ (a*) and ‘reaction involved in WNT signaling pathway’ classes (b*), thus it inherits the component_process_of ‘WNT_signaling pathway’ property (b**).
In the original paper, AlzPathway is described as a resource containing reactions and their corresponding pathways. Nevertheless, in AlzPathway, the reactions are not formally linked to corresponding pathways because the pathways are described as free text. We created classes corresponding to pathways by transforming the free text information into formal classes. Thus, we manually created classes corresponding to these pathways. Then, they were automatically linked to relevant reactions using description logic: for each pathway class, a class ‘reaction involved in pathway x’ was created and defined both as a ‘reaction that has_participant the molecules of interest in x’ and as a ‘component_process_of pathway x’. For example, the class ‘reaction involved in WNT signaling pathway’ has_participant ‘WNT’ and is a component_process_of ‘WNT signaling pathway’. Then, using automatic reasoning, all reactions having participants involved in pathway x were classified as subclasses of the ‘component_process_of pathway x’ classes and were linked to the pathway by subsumption with the component_process_of property. For example, ‘SFRP-WNT association’ is automatically classified as subclass of ‘reaction involved in WNT signaling pathway’ (see Figure 3b*) and inherits from its property: component_ process_of ‘WNT signaling pathway’ (see Figure 3b**). The 355 inferred SubClassOf axioms corresponding to reactions involved in one of the 22 pathways were added to the ontology. This resulted in an extended version of DMO, specific to AD physiopathology: ADMO.
Finally, in order to catch Single Nucleotide Polymorphisms (SNPs) measurements, we also added 7523 classes corresponding to SNPs from the NeuroChip SNP microarray (37), which are related to genes described in AlzPathway.
Results: ADMO content
Building ADMO resulted in a consistent network containing 2132 classes (1175 disjoint participants, including 88 phenotypes or signals, 1038 reactions and 22 pathways) linked with 10 964 logical axioms before and 12 373 logical axioms after automatic reasoning. The conversion of AlzPathway benefited from the DMO simple pattern of relationships (Figure 4A). Specific efforts were dedicated to the formal definition of the network with description logic axioms, leading to explicit relationships between processes, biological entities and pathways. These axiomatic definitions resulted in an increase of formalism compared to the initial representation of AlzPathway information. Following automatic reasoning, only 15 out of 643 AlzPathway’s reactions generically considered as ‘transition’ or ‘unknown transition’ remained unassigned to a specific process of the DMO upper model (e.g. ‘metabolic reaction’, ‘phosphorylation’ or ‘activation’). Moreover, 41 processes in AlzPathway were consistently assigned to a specific process different from their initial consideration (such as translation instead of transcription) and were, therefore, manually corrected. In addition, 355 reactions were formally defined as subprocesses of pathways thanks to automatic reasoning.
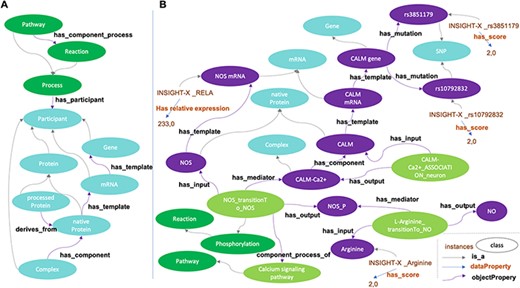
Disease Map Ontology (DMO) pattern (A) and application to Alzheimer Disease Map Ontology (ADMO) (B). AlzPathway derived-classes (B; illustrated for the Nitric Oxide Synthase phosphorylation and NO production) are now subclasses of DMO classes (A). Each class of ADMO may be instantiated by the corresponding entities as individuals. As illustrated in B, for a subject, scores for SNP rs3851179, RELA mRNA expression and Arginine measurement are linked by biochemical reactions.
Connection with ADO and use for storage of biomedical data: ADMO-plus
Mapping of ADMO with ADO
ADMO is a formal representation of AD pathophysiology at the molecular scale. It was designed to store and link omics biomedical data. Nevertheless, it would be interesting to also link data from other scales such as brain imaging or clinical scores (Figure 1C). ADO (29) describes knowledge not only about molecular processes (as in AlzPathway) but also about clinical assessments. By converting and integrating AlzPathway in OWL format, the resulting ontology and ADO are represented in the same format and thus can be connected with each other. In the first step, we selected ADO classes that correspond to ADMO ones. ADO classes were imported into ADMO independently of their initial hierarchy. Then, they were defined either (i) as equivalent classes of ADMO ‘process’, ‘pathway’, ‘phenotype’ or ‘gene’ classes (e.g. ADO: ‘Abeta-RAGE interaction’ class is equivalent to ADMO: ‘AB-RAGE_complexation’ class) or (ii) with DMO relationships towards ADMO classes (e.g. ADO: ‘macrophage activation’ class is equivalent to has_output ADMO: ‘activated microglia’ class or ADO: ‘neuron process’ class is equivalent to occurs_in ADMO: ‘neuron’ or ‘neuron compartment’ classes). Thus, for equivalent classes, ADO imported classes inherited from ADMO definition (e.g. ADO: ‘Abeta RAGE interaction’ class inherits the ADMO ‘AB-RAGE_complexation’ class definition: ‘protein–protein complexation’ that has for input ADMO: ‘RAGE’ and ADMO: ‘Amyloid decamere’ and has for output ADMO: ‘AB-RAGE’). For newly defined classes, automatic reasoning made it possible to build a new hierarchy between ADO and ADMO classes. All in all, 32 ADO classes were imported into ADMO (Supplementary Table S1-ADO) resulting in ADMO-plus.
Biomedical data integration
Ontologies’ classes can be filled by representative individual instances (a task called ‘instantiation’), which allows them to be used as resources for data storage. Thus, the next step consisted in instantiating ADMO-plus with biomedical omics data. As a proof of principle, SNP, gene expression (transcriptomic) and metabolomic data from the INSIGHT-PreAD study were used as instances to fill ADMO-plus classes (Figure 4B). The INSIGHT-PreAD is an ongoing prospective monocentric cohort with the objective to determine factors that increase the risk of progression of cognitively normal old adults to clinical AD. The study was approved by the local ethical committee (ANSM 130134B-31), and all participants signed a written informed consent. More information on the study is available in Supplementary text 1. As a proof of concept, we selected INSIGHT-PreAD genotypic, transcriptomic and metabolomic data that presented significant score variation. Among these data, only 16 SNPs corresponding to 11 genes (out of 53 SNPs corresponding to 44 genes), 23 mRNA relative expression (out of 145) and 25 metabolomic data (out of 53) could be integrated as classes’ instances in ADMO-plus. They were typed by their corresponding classes (for instance SNP, RNA or metabolite, see Figure 4B) and inherited from classes’ properties and thus from reaction and pathway information contained in the ADMO-plus network.
Discussion
We proposed the DMO ontological upper model in order to drive the conversion and integration of DMs into formal ontologies. We demonstrated its utility by converting AlzPathway (10) into an ontological model, called ADMO. It provides an increase in formalism, makes it interoperable with other ontologies (such as ADO (29), GO (26), the Protein Ontology (38)) and makes it able to integrate biomedical data. Based on a systems biology paradigm (39), all ADMO entities are formally defined as classes and interconnected within a consistent network. While AlzPathway contained several ambiguities, our efforts on formalism using description logic in the definition of ADMO classes allowed us to solve inconsistencies and provide a precise specification of processes and biological entities within the system. To our knowledge, there is no previous work on DM conversion to OWL, and this idea of converting an existing resource into an ontology is new.
Formalization of the fine description provided by DM
The increased formalism requires to assert a participant as a subclass of the most representative class and thus clarifies the status of the entities. In several standard bioinformatics knowledge resources (e.g. UnitProt (34), KEGG (35)), a same ID refers to a gene or a protein and in fine to a set of information, such as interactions, regulations and post-translation modifications (PTM), which are thus not specifically discriminated. However, omics technologies are able to generate data focused on specific elements of the systems (gene mutation, relative gene expression, protein concentration, ubiquitination ratio, phosophorylation ratio, etc.). When compared to other graph resources (14, 18, 40) that focus on genes and reactions only, DMs take each part of the system into consideration from genes expression to PTM (1). Our ADMO proposition goes one step further by formally defining the different elements of the system. By providing disjoint classes for different molecular states, DMO breaks ambiguities between genes, genes product and their modified states (27). ADMO can be instantiated with omics data within the specific corresponding classes, resulting in an ontology that explicitly integrates each type of omics data despite the complexity of the AD pathophysiology system.
Automatic reasoning facilitated by the systems paradigm
Taking advantage of a systems biology approach and reasoning properties, DMO can automatically ensure satisfiability of ADMO and provides inferences of hierarchy and new relationships (such as the link between a pathway and its process components) (25). Other ontologies provide generic models in the field of molecular biology, such as BioPax (41). BioPax is a well-established framework to share information between knowledge bases. However, it was essentially designed to manage knowledge sharing, and logical reasoning is limited to satisfiability check. While DMO presents a level of genericity similar to that of BioPax, it is designed to manage subclasses (e.g. ADMO) that in turn manage data as their instances. Moreover, due to its management by automatic logical reasoning and its model based on systems paradigm, ADMO is particularly flexible. If reaction, participant or pathway classes are removed or added, automatic reasoning is able to rebuild the modified network in a consistent way. In addition, class instantiation facilitates this task and instanced data take advantage of an up-to-date network. In existing resources, biological entities are mainly annotated by the cellular component, molecular function or biological process classes from GO (42). While this provides the largest set of data indexed by a controlled vocabulary (26), this is also limited by the fact that genes are not gene products, nor functions or processes. Thus, genes cannot be assigned as individual instances of GO classes, leading to an underuse of the reasoning ability provided by ontologies. With the classical annotation methods, a knowledge change, such as the addition or the removal of a new molecular reaction, involves a verification of all annotated entities that were implicitly related to the process concerned. With instantiation methods, relationships between participants and processes are explicitly expressed as axioms and a modification in the ontology directly applies to instanced entities.
Finally, automatic reasoning also provides formal relationships between reactions and their related pathways, which did not exist in AlzPathway. The formal specification of pathways and their relationships with reactions in ADMO opens new promises for mapping ADMO with widely used ontologies such as GO or the Human Phenotype Ontology (43).
Connection with other resources and potential extensions
Converted in OWL format, AlzPathway’s knowledge is now interoperable with other ontologies such as ADO (29). This results in a mutual enrichment: ADMO is linked with clinical knowledge and ADO benefits from more specific knowledge about AD pathophysiology. Here, we present a first attempt to connect ADMO/ADO into ADMO-plus, as a proof of concept. Going further would necessitate a revision of ADO, which is designed for text mining and which is not adapted to logical reasoning and systems biology. The OWL format also opens perspectives to integrate other knowledge resources. Here, we relied on AlzPathway, but additional resources could be used. In the domain of AD, the knowledge graph neuroRDF (44) would increase current knowledge provided by AlzPathway and ADO. Our DMO upper ontological model also provides an interesting framework to embed generic resources and thus harmonize AlzPathway and those resources. This offers new avenues for increasing the scale of representation of AD pathophysiology network in our framework. Indeed, our approach shall facilitate the future update of AlzPathway. Ontologies manage knowledge in a network as a directed graph. Thus, when reactions are added or removed, they are automatically integrated within or suppressed from the network. As ADMO is based on automatic reasoning for classification, any new reaction described with its reactants will automatically be described hierarchically by its type and linked to a previously described pathway. Nevertheless, the integration of biomedical data from the INSIGHT-PreAD study underlined the fact that the range of data identified as significant in this study is larger than the coverage of ADMO-plus (which reflects the current knowledge specific to AD pathophysiology). While AlzPathway currently provides the larger network in the domain, it is a compilation of knowledge from a reductionism approach. This suggests that the network has to be extended in order to generate new hypotheses about AD etiology (13).
Our strategy could be applied to other DMs and increase their interoperability. The main limitation is the availability of DM of interests. Apart from that, we believe that this work can be applied to other DMs since DMO was designed to embed any DM and can easily be reused. In practice, there will be specific parts that will need to be adapted, specifically the extraction of the DM content and its possible modifications made by a domain expert. Then, other tasks can be easily done using the Protégé editor.
Even though they are less specific than DM, considering generic systems resources, such as Reactome (2), would provide useful additional information through a wide range of generic curated biochemical reactions and pathways. The integration, in the same framework, of disease-specific and non-specific pathways is useful to build a more comprehensive view of the disease, in particular if these pathways are interconnected. Then such integration is done, OWL format allows to tag pathways as disease specific or not, so that the user can be clear about their status, leading to an enrichment without adding noise. In the same way, the genericity of processes and participants described in the DMO upper model opens the perspective to harmonize specific DM with an equivalent curation level from other neurodegenerative disorders, such as the Parkinson’s DM (3).
Differences with DM
Unlike DM (1), ontologies are not adapted to graphical visualization (45). Thus, DMs are better adapted to human reading. On the other hand, ontologies can store the network (classes and relationships) in a more consistent way and can thus be tested for logical consistency. Also, they present a higher flexibility to formally integrate new elements in the knowledge graph, as we did by adding 865 genes, related SNPs and mRNAs. Note that, during the conversion step, AlzPathway’s internal IDs were retained as class annotations, allowing interoperability and retrieval between the initial (graphical) and converted (formal) resource. Thus, DM and ontologies are complementary approaches. The combination of these two approaches will be beneficial for both the DM and bio-ontology communities.
In conclusion, we proposed a generic approach to transform DMs into formal ontologies. We demonstrated its use through the conversion of an Alzheimer’s DM. This enriches it with reasoning properties and makes it interoperable with other ontologies. This should constitute a useful resource for the community. The approach is generic and can be applied to other DMs.
Supplementary data
Supplementary data are available at Database Online.
Funding
The research leading to these results has received funding from the French government under the management of Agence Nationale de la Recherche as part of the ‘Investissements d’avenir’ program, reference ANR-19-P3IA-0001 (PRAIRIE 3IA Institute) and reference ANR-10-IAIHU-06 (Agence Nationale de la Recherche-10-IA Institut Hospitalo-Universitaire-6), from the Inria Project Lab Program (project Neuromarkers) and from the Fondation Vaincre Alzheimer (grant number FR-18006CB).
The sponsors had no role in the study design, data analysis or interpretation, writing or decision to submit the report for publication.
Author contributions
Dr Vincent Henry had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concepts and study design: V.H., I.M., O.C.
Acquisition, analysis or interpretation of data interpretation: all authors.
Manuscript drafting or manuscript revision for important intellectual content: all authors.
Approval of final version of submitted manuscript: all authors.
Literature research: V.H.
Obtained funding: O.C., I.M., V.H.
Study supervision: O.C., I.M.
Conflict of interest.
None declared.
References
Appendix 1. List of abbreviations
AB/Abeta: Amyloid β
AD: Alzheimer’s disease
ADMO: Alzheimer’s Disease Map Ontology
ADO: Alzheimer’s Disease Ontology
APOE: Apolipoprotein E
DM: Systems medicine disease maps
DMO: Disease Map Ontology
GO: Gene ontology
HPO: Human Phenotype Ontology
KEGG: Kyoto Encyclopedia of Genes and Genomes
mEPN: modified Edinburg Pathway Notation
(m)RNA: messenger ribonucleic acid
OWL: Web Ontology Language
PTM: post-translation modifications
RAGE: receptor for advanced glycation endproducts
RO: Relation ontology
SBML: Systems Biology Markup Language
SBO: Systems Biology Ontology
SFRP: Secreted frizzled-related protein
SNP: single-nucleotide polymorphism
WNT: Proto-oncogene protein Wnt
Appendix 2. Glossary
Accuracy: measures how close the measurements are to a specific value.
Cardinality: the cardinality between two sets is the numerical constraint between individual instances of one set and individual instances of the other.
Class disjunction: logical property which formally separates two (or more) classes. It means that if an individual is a member of a class, it cannot be member of the other classes.
Existential restriction: an existential quantification is a type of quantifier which is interpreted as ‘there exists’, ‘there is at least one’, or ‘for some’.
Expressiveness: the expressive power of a language is the breadth of ideas that can be represented and communicated in that language. The more expressive a language is, the greater the variety and quantity of ideas it can be used to represent.
Relationships: relationships (also known as relations) between objects in an ontology specify how objects are related to other objects. Typically, a relation is of a particular type (or class) that specifies in what sense the object is related to the other object in the ontology.
Logical axioms: assertions (including rules) in a logical form that, together, comprise the overall theory that the ontology describes in its domain of application.
Types (i.e. the classes): collection of sets that can be unambiguously defined by a property that all its members share.
Universality: a universal quantification is a type of quantifier which is interpreted as ‘given any’ or ‘for all’. It expresses that a propositional function is satisfied by every member of a domain of discourse.



