-
PDF
- Split View
-
Views
-
Cite
Cite
Nan Deng, Canglin Wu, Ashraf Yaseen, Hulin Wu, ImmuneData: an integrated data discovery system for immunology data repositories, Database, Volume 2022, 2022, baac003, https://doi.org/10.1093/database/baac003
Close - Share Icon Share
Abstract
To meet the increasing demand for data sharing, data reuse and meta-analysis in the immunology research community, we have developed the data discovery system ImmuneData. The system provides integrated access to five immunology data repositories funded by the National Institute of Allergy and Infectious Diseases, Division of Allergy, Immunology and Transplantation, including ImmPort, ImmuneSpace, ITN TrialShare, ImmGen and IEDB. ImmuneData restructures the data repositories’ metadata into a uniform schema using domain experts’ knowledge and state-of-the-art Natural Language Processing (NLP) technologies. It comes with a user-friendly web interface, accessible at http://www.immunedata.org/, and a Google-like search engine for biological researchers to find and access data easily. The vast quantity of synonyms used in biomedical research increase the likelihood of incomplete search results. Thus, our search engine converts queries submitted by users into ontology terms, which are then expended by NLP technologies to ensure that the search results will include all synonyms for a particular concept. The system also includes an advanced search function to build customized queries to meet higher-level users’ needs. ImmuneData ensures the FAIR principle (Findability, Accessibility, Interoperability and Reusability) of the five data repositories to benefit data reuse in the immunology research community. The data pipeline constructing our system can be extended to other data repositories to build a more comprehensive biological data discovery system.
Introduction
In the era of big data, the exponential growth of biological data, especially publicly available data, produces ample opportunities for biological research and some challenges for the researchers. They can reuse public data to raise and investigate further biological questions or evaluate current methods’ repeatability without spending an enormous amount of time and money in generating data. Thus, data should be managed in a findable, accessible, interoperable and reusable manner. These criteria are called FAIR (Findability, Accessibility, Interoperability and Reusability) guiding principles (1). Several large immunological data repositories are funded by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID), Division of Allergy, Immunology and Transplantation to promote the reuse of data. However, there is still room to improve the FAIR-ness of those data repositories. Specifically, an index-and-search system that grants access to those data resources can enhance the findability and accessibility of data.
In this work, we have integrated five data repositories funded by NIAID, shown in Table 1. ImmPort (2, 3) is a data repository that contains data from NIAID-funded immunology studies, including basic research and clinical trials. ImmuneSpace (4) is a data management and analysis platform that mainly provides data from the Human Immunology Project Consortium. ITN TrialShare (5) is a clinical trial data portal for the Immune Tolerance Network (ITN). It shares information about the ITN’s clinical studies and specimens, as well as data and analysis results underlying ITN’s publications. ImmGen (6) hosts microarray data from the immune system of mice funded by the Immunological Genome Project. IEDB (7, 8) is a knowledge-based resource (http://www.iedb.org) allowing users to search antibody and T-cell epitopes related to experimental data in different species, such as humans, non-human primates and other animal species. Epitopes involved in infectious disease, allergy, autoimmunity and transplant are included. It also hosts analysis tools for B-cell and T-cell epitopes. Unlike the other four data repositories, this repository does not store the raw data but the ‘conclusions’ of epitopes with the references. Thus, we define those data as ‘knowledge.’
| . | Type . | Number of datasets . | Clinical researches . | Basic research . | Species . |
|---|---|---|---|---|---|
| ImmPort | Data | 301 | Yes | Yes | Multiple |
| ImmuneSpace | Data | 74 | Yes | Yes | Homo sapiens |
| ITN TrialShare | Data | 28 | Yes | Homo sapiens | |
| ImmGen | Data | 865 | Yes | Mus musculus | |
| IEDB | Knowledge | 1 509 085 | Yes | Yes | Multiple |
| . | Type . | Number of datasets . | Clinical researches . | Basic research . | Species . |
|---|---|---|---|---|---|
| ImmPort | Data | 301 | Yes | Yes | Multiple |
| ImmuneSpace | Data | 74 | Yes | Yes | Homo sapiens |
| ITN TrialShare | Data | 28 | Yes | Homo sapiens | |
| ImmGen | Data | 865 | Yes | Mus musculus | |
| IEDB | Knowledge | 1 509 085 | Yes | Yes | Multiple |
| . | Type . | Number of datasets . | Clinical researches . | Basic research . | Species . |
|---|---|---|---|---|---|
| ImmPort | Data | 301 | Yes | Yes | Multiple |
| ImmuneSpace | Data | 74 | Yes | Yes | Homo sapiens |
| ITN TrialShare | Data | 28 | Yes | Homo sapiens | |
| ImmGen | Data | 865 | Yes | Mus musculus | |
| IEDB | Knowledge | 1 509 085 | Yes | Yes | Multiple |
| . | Type . | Number of datasets . | Clinical researches . | Basic research . | Species . |
|---|---|---|---|---|---|
| ImmPort | Data | 301 | Yes | Yes | Multiple |
| ImmuneSpace | Data | 74 | Yes | Yes | Homo sapiens |
| ITN TrialShare | Data | 28 | Yes | Homo sapiens | |
| ImmGen | Data | 865 | Yes | Mus musculus | |
| IEDB | Knowledge | 1 509 085 | Yes | Yes | Multiple |
The data repositories are well designed, but data are stored in substantially diverse structures. Researchers inevitably need to search for different types of data crossing multiple data repositories. Without an integrated index and search system, this process would be time-consuming and frustrating. Furthermore, a user might only be familiar with a few but not all of these data repositories, so that some valuable datasets may be neglected. Thus, an integrated data discovery system containing these data repositories is desired to improve the findability and accessibility of available public immunological data.
One significant challenge in data integration is the heterogeneity of the metadata. To solve this problem, we adopted an existing unified metadata model, the DatA Tag Suite (DATS) model (9). DATS model is a generic and platform-independent model designed to store various types of biomedical data. This model also facilitates effective searching and comparison across multiple data repositories. In the future, this unified DATS model will ensure the expandability of our system and simplify the data sharing process with other databases or search engines.
The DATS model we used in our system is DataMed (9, 10). We retrieved 1268 datasets and 1 509 085 ‘knowledge’ records from the five data repositories, as shown in Table 1. After consultation with domain experts and Natural Language Processing (NLP) enhancement of the DATS model adoption and metadata, we produced a database with a unified metadata format.
We developed a user-friendly web interface with an Elasticsearch-powered engine at (https://immunedata.org/) so that immunologists can utilize our website without difficulty. Immunedata website provides a one-stop, all-inclusive solution for data seekers that is time-efficient and easily accessible in the field of Immunology. It will significantly promote the efficiency of data searching and the capability to reuse public data in this scientific community.
Methods and data description
Data source
Five different immunology databases are included in the data discovery system: ImmPort (2), ImmuneSpace (4), ITN TrialShare (5), IEDB (7) and ImmGen (6). IEDB’s metadata has been directly downloaded from the official website. ImmGen’s metadata is deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/). However, ImmPort, ImmuneSpace and ITN TrialShare provide neither data download function nor any API. We created web crawlers for these three databases using Python programming language.
Data representation
Metadata from different databases are in heterogeneous schemas so that standardization of metadata is required to improve these databases’ interoperability. We utilized and customized the extended elements of DATS to accommodate immunology data while keeping the generic core elements applicable to any data type. Domain experts inspected existing metadata defined in each resource to figure out their overlap, uniqueness and supported use cases. The principal inclusion criteria of existing ontologies and data standards are based on the importance of the terms in the immunology research (e.g. data sharing, knowledge dissemination, standard development and integrative analyses).
The schema of DATS model can be obtained from bioCADDIE project (9) at https://biocaddie.org/group/working-group/working-group-3-descriptive-metadata-datasets. In the record detail pages, more information is listed in the DATS structure such as title, dataset information (id, description, etc.), accessibility (link, authorizations, etc.), dimension (species, conditions, study type, etc.), disease, author, study group, original data repositories and more. Such information helps users to identify and download the data they need.
Microservices
We use microservice architecture to implement our essential functions. With microservices, the application can be divided into small components, independent from each other. Instead of a traditional, monolithic approach, where an application is built in a single large construction, multiple components separately perform various tasks and then coupled into one application. Each of these components is a microservice. We have deployed and optimized NLP and terminology microservices on AWS docker instances. A testing service was also built to validate the performance of the application. The system microservice architecture is shown in Figure 1.
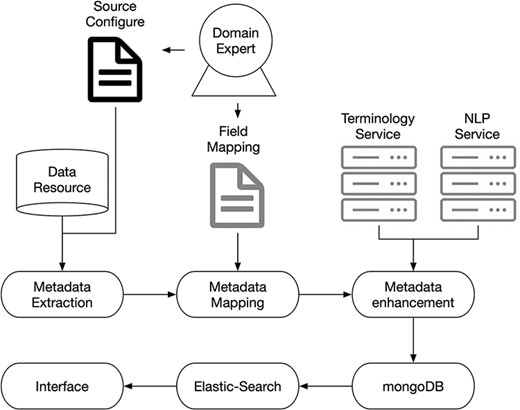
Under the guidance of domain experts, we decided on our data schema and field mapping strategy. The metadata was exacted from data resources and mapped into a uniform schema according to the mapping strategy. Then, terminology and NLP services were involved in enhancing the metadata. After that, the enhanced metadata were stored in a MongoDB server. Finally, the elastic-search powered interface was used to provide the searching function to the end users.
Google-like searching
After a user submits a query, biomedical concepts are extracted by the NLP service. Multiple major ontologies or controlled vocabulary thesaurus, including MeSH (https://www.ncbi.nlm.nih.gov/mesh) (11), SNOMED CT (http://www.snomed.org/) (12), Gene Ontology (https://www.geneontology.org/) (13), Foundational Model of Anatomy (https://si.washington.edu/projects/fma) (14), NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy) (15) and Hugo Gene Nomenclature (https://www.genenames.org/) (16), are used in our terminology service to expand the user’s query to a list of synonyms to improve query coverage. The terminology service is based on SciGraph (https://www.springernature.com/gp/researchers/scigraph) and Neo4j (http://neo4j.com/). In our search engine, Elasticsearch (https://www.elastic.co/) serves as the core to perform the searching function. Elasticsearch is the most popular enterprise search engine based on Lucene (https://lucene.apache.org/). It is a distributed, multitenant-capable full-text search engine with schema-free JSON documents. All the metadata from the ingestion pipeline was indexed in the Elasticsearch endpoint and connected to the user interface.
Web interface
We have established a user-friendly PHP-based web application by following the Model-View-Controller pattern. Our interface provides search results and faceted navigation, which help end-users filter and refine results. Details of the interface will be described in the ‘Results and Discussions’ section.
Results and discussions
ImmuneData: an overview
Currently, our database contains datasets from five data repositories. Those datasets can be grouped into two categories based on the data type they store. One category is ‘experimental data’, which contains raw data from immunology assays or tests. We were able to catalog 1268 datasets of this type. The other category is ‘knowledge’ data. Each knowledge entry is either a conclusion from biological experiments or information about how this conclusion is reached, i.e. reference and metadata of bioassays. Raw data is not included in these entries. For this category, we recorded 1 509 085 knowledge entries.
Searching function
In the biology domain, a term may have many different synonyms. For example, flu was also known as influenza and was called ‘grippe’ in the old days. Thus, in addition to the user’s keywords, we also use their synonyms to perform the query. For example, when searching for influenza, we build the query (data) ‘flu’ OR [(‘Influenzas’ OR ‘Influenza Human’ OR ‘Human Influenzas’ OR ‘Influenza (disorder)’ OR ‘Influenzas, Human’ OR ‘Human Influenza’ OR ‘Flu’ OR ‘Human Flu’ OR ‘Influenza Humans’ OR ‘Influenza, Human’ OR ‘Grippe’ OR ‘Flu, Human’)]. This search strategy improves the coverage of the results. This synonym search function can be disabled by quoting the keywords, i.e. when using ‘flu’ as the keyword between quotation marks, influenza and grippe will not be included in the query. Searching with ‘flu’ only returned 460 results, and 58 of them are datasets, while searching with influenza (without quoting) returned 6520 results, and 98 of them are datasets. For example, in the dataset ‘Vaccination with drifted variants of H5 hemagglutinin protein elicits a broadened antibody response’, the author used influenza but not flu in the description, which can only be found by the synonym search. To make sure it does not compromise search results’ precision, we tested the search function with 10 important disease names (Table 2) as keywords. There is no evidence showing that the search’s precision was compromised, and the overall precision is 96.1% (123/128) in those 10 search results. The only term with a low precision is related to ‘AIDS’, which returns studies such as ‘memory aids’. This kind of false-positive is hard to eliminate.
| . | Precision . | Retrieved datasets . | Correct . |
|---|---|---|---|
| Lupus | 1 | 11 | 11 |
| Rheumatoid arthritis | 0.86 | 7 | 6 |
| Multiple sclerosis | 1 | 9 | 9 |
| Allergy | 0.86 | 14 | 12 |
| HBV | 1 | 5 | 5 |
| AIDS | 0 | 2 | 0 |
| Asthma | 1 | 15 | 15 |
| Flu | 1 | 51 | 51 |
| Tuberculosis | 1 | 5 | 5 |
| Cancer | 1 | 9 | 9 |
| Total | 0.96 | 128 | 123 |
| . | Precision . | Retrieved datasets . | Correct . |
|---|---|---|---|
| Lupus | 1 | 11 | 11 |
| Rheumatoid arthritis | 0.86 | 7 | 6 |
| Multiple sclerosis | 1 | 9 | 9 |
| Allergy | 0.86 | 14 | 12 |
| HBV | 1 | 5 | 5 |
| AIDS | 0 | 2 | 0 |
| Asthma | 1 | 15 | 15 |
| Flu | 1 | 51 | 51 |
| Tuberculosis | 1 | 5 | 5 |
| Cancer | 1 | 9 | 9 |
| Total | 0.96 | 128 | 123 |
| . | Precision . | Retrieved datasets . | Correct . |
|---|---|---|---|
| Lupus | 1 | 11 | 11 |
| Rheumatoid arthritis | 0.86 | 7 | 6 |
| Multiple sclerosis | 1 | 9 | 9 |
| Allergy | 0.86 | 14 | 12 |
| HBV | 1 | 5 | 5 |
| AIDS | 0 | 2 | 0 |
| Asthma | 1 | 15 | 15 |
| Flu | 1 | 51 | 51 |
| Tuberculosis | 1 | 5 | 5 |
| Cancer | 1 | 9 | 9 |
| Total | 0.96 | 128 | 123 |
| . | Precision . | Retrieved datasets . | Correct . |
|---|---|---|---|
| Lupus | 1 | 11 | 11 |
| Rheumatoid arthritis | 0.86 | 7 | 6 |
| Multiple sclerosis | 1 | 9 | 9 |
| Allergy | 0.86 | 14 | 12 |
| HBV | 1 | 5 | 5 |
| AIDS | 0 | 2 | 0 |
| Asthma | 1 | 15 | 15 |
| Flu | 1 | 51 | 51 |
| Tuberculosis | 1 | 5 | 5 |
| Cancer | 1 | 9 | 9 |
| Total | 0.96 | 128 | 123 |
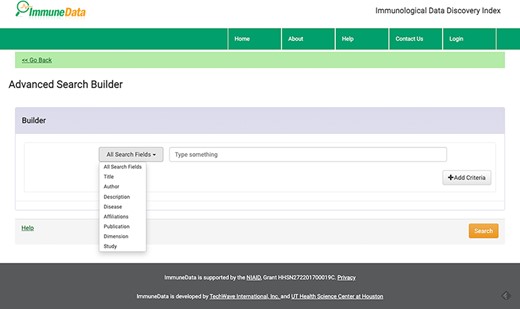
We offer two search modes: standard Google-like search and advanced search to accommodate different levels of users. In our system, rich information is provided for each returned result. For example, when we conduct a basic search using the keyword ‘influenza’, 31 642 results were returned, as seen in Figure 2. Using the advanced search function, one can build a customized search with various fields, such as Title, Author, Description, Disease, Affiliation, Publication, Dimension and Study, as you can see in Figure 4. For example, we can search for influenza in the disease field and NIAID in the affiliation field and get two datasets about influenza generated by NIAID.
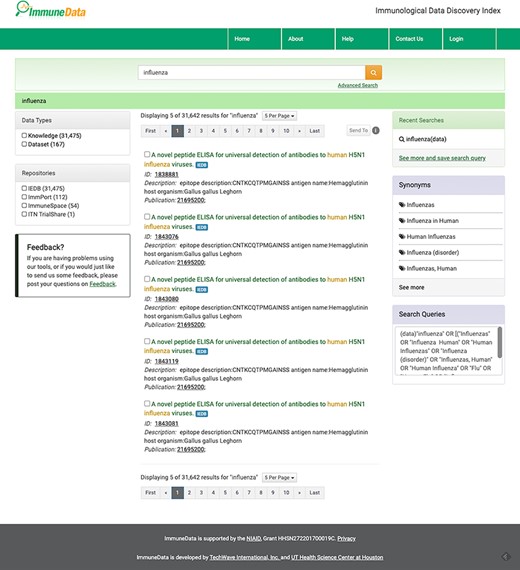
The result page of basic search using ‘influenza’ as the keyword.
When a user is interested in information on a specific epitope, they can directly search for its amino acid sequence. For example, if we search for SIINFEKL, a CD8 epitope in the influenza virus, 688 knowledge records describing this epitope were returned.
In the search result page in Figure 3, the title, ID, description and related reference’s PubMed ID are listed for each result. In the left panel of the search results page, a user can select the data type and the data source of those results. On the right panel of the page, the recent search history is listed at the top. Additionally, synonyms used in the search and the queries command are also listed in the right panel.
Conclusion and future directions
In summary, we established a comprehensive database system containing several major immunology data repositories. It features a unified DAT format of metadata and an optimized search function, making the database user-friendly, especially to experimental biologists who may not have advanced computer skills. It will encourage the reuse of data in the immunology research community. The data integration standards, technologies and tools in this system can also be used in other data repositories. In the future, we will integrate more immunology data repositories into the ImmuneData system, as well as extend our pipeline to other data repositories in different biomedical sub-domains and form a universal data discovery system for the whole biological research community.
Acknowledgements
We thank Dr Hua Xu, Dr Xiaoling Chen and Dr Ergin Soyal for assistance with DATS model. We would also like to show our gratitude to Isabella Wu for proofreading.
Funding
National Institutes of Health Grant HHSN272201700019C, P30 AI161943, CPRIT RP170668.
Conflict of interest
None declared.
References
(



