-
PDF
- Split View
-
Views
-
Cite
Cite
Baiyang Feng, Jing Gao, AnthraxKP: a knowledge graph-based, Anthrax Knowledge Portal mined from biomedical literature, Database, Volume 2022, 2022, baac037, https://doi.org/10.1093/database/baac037
Close - Share Icon Share
Abstract
Anthrax is a zoonotic infectious disease caused by Bacillus anthracis (anthrax bacterium) that affects not only domestic and wild animals worldwide but also human health. As the study develops in-depth, a large quantity of related biomedical publications emerge. Acquiring knowledge from the literature is essential for gaining insight into anthrax etiology, diagnosis, treatment and research. In this study, we used a set of text mining tools to identify nearly 14 000 entities of 29 categories, such as genes, diseases, chemicals, species, vaccines and proteins, from nearly 8000 anthrax biomedical literature and extracted 281 categories of association relationships among the entities. We curated Anthrax-related Entities Dictionary and Anthrax Ontology. We formed Anthrax Knowledge Graph (AnthraxKG) containing more than 6000 nodes, 6000 edges and 32 000 properties. An interactive visualized Anthrax Knowledge Portal(AnthraxKP) was also developed based on AnthraxKG by using Web technology. AnthraxKP in this study provides rich and authentic relevant knowledge in many forms, which can help researchers carry out research more efficiently.
Database URL: AnthraxKP is permitted users to query and download data at http://139.224.212.120:18095/.
Introduction
Anthrax, also known as Woolsorter’s disease (1), is an acute infectious disease caused by the bacterium Bacillus anthracis that often occurs in cattle, sheep, goats, horses, which are herbivorous livestock and other wildlife (2). Anthrax can be transmitted to people mainly through cutaneous, respiratory, gastrointestinal transmission and injection (3). Cutaneous infection is the most common way of human infection, accounting for >95% of total cases, and inhalation anthrax is the most lethal, with a mortality rate of >85% if left untreated (4). Anthrax has the properties of a high lethality rate, multiple routes of infection and easily cultured spores, so anthrax spores have been developed as a biological weapon by many countries (5, 6). Intensive anthrax research efforts have led to an increasing number of scientific publications, and nearly 8000 anthrax-related literature exists on PubMed, but by far there is no biomedical ontology database for anthrax.
In this study, we constructed Anthrax-related Entities Dictionary (AED) and Anthrax Ontology (AO). In the biomedical field, ontology knowledge bases, including Gene Ontology (GO) (7), Disease Ontology (DO) (8), Protein Ontology (PRO) (9) and Human Phenotype Ontology (HPO) (10), can provide high-quality data sources for researchers. However, due to the wide range of knowledge covered by biomedical texts and the high learning cost, there has been a large amount of unstructured knowledge to be mined in the relevant literature (11), and the previously constructed knowledge bases are limited to the level of few entity categories and single association relationships, such as ontology structure of the gene–phenotype relationship (12, 13) and the gene–disease association (14, 15). The purpose of this research is to curate a robust AO knowledge base to host 281 association relationships among 29 categories of entities.
We used a semi-automatic extraction method (16, 17) to construct AO. First, we use an automated script to segment the text and retain the statements where knowledge is likely to occur, then we automatically extract the knowledge through natural language processing tools and finally, we correct some irrelevant results through manual verification. Early biomedical knowledgebases were curated and maintained manually, as the manually extracted knowledge usually had a high accuracy rate (18). The Catalogue of Somatic Mutations in Cancer (COSMIC) (19), which is manually curated by experts, is a typical example. However, under the premise of a high publication rate of scientific literature (20), the manual methods have a low recall rate (21), but it is particularly important for establishing a standard data set, as well as evaluating and training models (18). Most automatic methods are based on natural language processing techniques (16). Universal protein resource (22) is curated automatically. This method needs good models and training data for support, but currently, the biomedical text corpus for anthrax has not been formed, and the automatic methods may ignore the implicit relationship in multiple statements (23). Our semi-automatic method largely reduces the number of text statements to be filtered, which saves a lot of time compared to the manual way (24), thus improving the recall rate, and the accuracy rate is improved much higher than that of the automatic way.
In this study, we constructed Anthrax Knowledge Graph (AnthraxKG). Before this, there was no such biomedical knowledge graph (KG) for anthrax in the field. We mapped the data of AO into AnthraxKG, which uses nodes to represent biomedical concepts and edges to represent association relationships, and carried various property information. KGs (25, 26) are knowledge bases that use graph structure data models or topology to integrate data (27), and many biomedical KGs have entered people’s vision, such as OpenKG (28), SemaTyP (29), OpenBiolink (30), protein-drug target KG (31) and the recently updated COVID-19 KG (32). This visual representation method applied in the biomedicine field can promote the efficient understanding of biomedical knowledge by scientific researchers, improve scientific research efficiency, boost scientific research ideas, and expand the scope of research, thus potentially promoting clinical decision-making and promoting precision medicine, it can help to discover new disease prevention and treatment methods (detection technology, chemicals, vaccines, treatment methods) (33, 34).
In order to enhance the operability and user-friendliness of the knowledge base, we used Web technology to build a managed, multi-module, interactive visualized, KG-based Anthrax Knowledge Portal (AnthraxKP), which allows researchers to query, browse and download anthrax biomedical resources in multiple formats.
Methods
At present, there is much-unstructured anthrax knowledge in scientific literature. This paper designed a set of process methods to extract knowledge from anthrax scientific literature. Search for ‘Anthrax’ from PubMed (https://pubmed.ncbi.nlm.nih.gov/) to get the abstracts of anthrax-related literature. Take the abstracts as the raw data, split them into preprocessed statements and use a set of tools and methods to identify entities and extract the relationship between entities. The ontology (schema layer) in the form of ‘entity-relationship-entity, entity-property-value’ is obtained. After manual inspection and correction of data, we use the graph database to store structured data to form AnthraxKG (data layer). Finally, AnthraxKP is developed through Web technology. The workflow is explained in Figure 1.
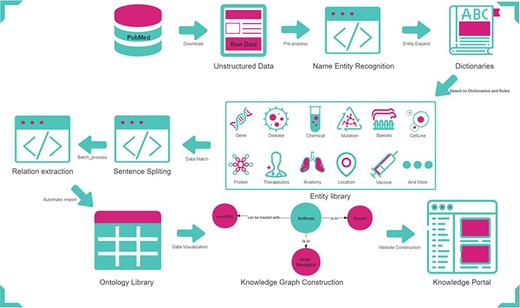
Data source and preprocessing
Search for ‘Anthrax’ on PubMed, and get the abstracts of 7764 anthrax-related literature. We use these abstracts as the data source. PubTator Central (35) provides automatic annotations of biomedical concepts, which can be accessed interactively through a Web browser, programmatically via RESTful API or batch download via File Transfer Protocol. We use this tool to annotate the entities in the abstracts and get the preprocessed abstracts in PubTator format.
Named entity recognition and construction of AED
PubTator-annotated biomedical concepts (PBCs) include Gene, Disease, Chemical, Species, DNAMutation, ProteinMutation, Single Nucleotide Polymorphism (SNP) and CellLine. To improve the coverage of biomedical concepts, we augmented these eight PBCs with 21 entity categories for AED by a dictionary-based approach combined with manual annotation, the 21 entity categories are also derived from the anthrax-related biomedical literature. Among them, Vaccine, Toxin, Amino Acid, Peptide, Protein, Nucleoside, Nucleic acid, Nucleotide and Enzyme are curated based on PBC_Chemical; Anatomy, Phenomena, Location, Equipment, Agency, Genome, Behavior, Therapeutics, Diagnosis, Technique, Person and Food are curated according to the actual needs of AO.
The main method refers to the National Center for Biotechnology Information (NCBI) (36) IDs (NCBI_IDs) of the PBC entities: Gene_ID, Species_ID, CVCL_ID, DNAMutation_ID, ProteinMutation_ID, SNP_ID, MeSH_ID to curate AED. Medical Subject Headings (MeSH) (37) are hierarchical organization terms used for indexing and cataloging biomedical information. It is used to index PubMed and other National Library of Medicine databases. The NCBI provides a complete MeSH data set and keeps it updated and maintained, and the entities with MeSH_ID as NCBI_ID have the largest proportion in AED, herein is used as an example: download the MeSH data set in XML format as a dictionary and convert it into JavaScript Object Notation (JSON) format through the xml2json tool (https://github.com/hay/xml2json). Then, according to the hierarchical relationship of MeSH thesaurus, select the subject words with high word frequency and apply them to AnthraxKG as the augmented entity categories, match the corresponding entities according to MeSH_ID, extract the corresponding synonyms (Entry_Terms) to curate the AED and finally correct the error information manually.
Note: The NCBI_ID categories are different, but the annotation methods are basically the same, so we will not repeat them here. The category of unannotated entities is defined as ‘Other’.
Relation extraction
The preprocessed sample for this study is complex, which belongs to a specialized and sophisticated academic field, and there is no existing biomedical text corpus for anthrax. We used the Open-domain information extraction system (OpenIE) (38) to extract relations. Stanford OpenIE is part of Stanford CoreNLP (39), a tool that extracts structured triples from text without specifying relations and training corpus in advance. First, we used Natural Language Toolkit (40) to split the literature abstracts into sentences. Then the sentences containing entities in the AED were retained using regular expression operations in Python (https://docs.python.org/3/library/re.html), totaling 37 433 sentences. Finally, we use OpenIE for relation extraction (RE), combined with manual curation to get structured triples. For example, the sentence ‘19 654 018|B. anthracis is the etiologic agent of anthrax.’ is processed to obtain a triple ‘|-{“subject”: “B. anthracis”, “relation”: “is etiologic agent of”, “object”: “anthrax”}’.
AO construction
Named entity recognition (NER) and RE tasks provide a large amount of structured data. We curated AO in Comma-Separated Values (CSV) format with a fixed conceptual framework (Entity1-Entity_Category-Entity_id-Relation-Entity2-Entity_Category-Entity_id-Source); meanwhile, we annotated the PubMed identifier (PMID) for researchers to trace and validate.
AnthraxKG construction
The Neo4j graph database management system can store structured knowledge and support semantic queries. We deployed Neo4j as the graph database of AnthraxKG, converted the AO in CSV format to JSON format by the csv2json tool (https://github.com/oplatek/csv2json) and embedded the AO data in the Neo4j graph database using the py2neo interface.
AnthraxKP construction
We have developed a multi-module database management system: AnthraxKP, which can be accessed at http://139.224.212.120:18095/. AnthraxKP is constructed using the Python Django Web framework for the front-end and implemented with Neo4j + MySQL for the back-end. The website consists of four modules as described below.
AnthraxKG visualization module
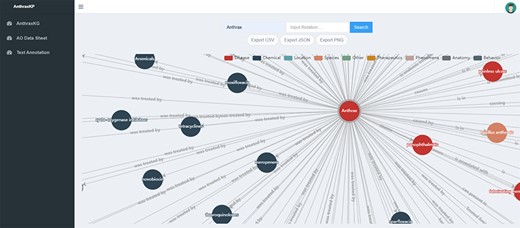
This module is the visualization area of AnthraxKG, as shown in Figure 2, and it can display nodes and edges, query on nodes and edges and traverse out hierarchical nodes and relationships, providing data export functions in CSV, JSON and Portable Network Graphics formats. Although Neo4j Browser allows users to interact with the graphs and visualize the information, it serves as a developer-centric tool that needs to perform Cypher queries. To facilitate users to interact with the data in a more user-friendly way, we set up a keyword search function. We used Pyecharts to render AnthraxKG visually, and the user does not interact directly with the database when accessing the front-end pages to improve the interaction rate.
AO datasheet module
In this module, users can browse the list of AO data, can search and filter the data and export the data in XLSX and CSV formats.
Automatic text annotation module
This module provides automatic annotations of 29 categories of entities and their corresponding relationships in the literature abstracts with different colors and displays the entity category and NCBI_ID. Users only need to enter PMID, article title or keywords in the search box to get a response.
Developer module
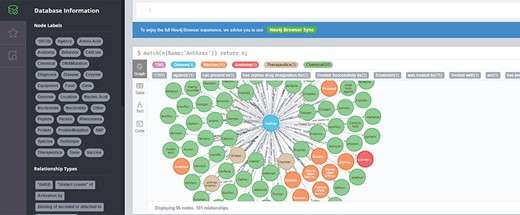
This module is mainly used by developers or domain experts to maintain data; the module supports adding, deleting, checking and correcting AO data. It also supports access to the backend data through the Neo4j Browser, as shown in Figure 3. Developers can update AO and anthrax scientific literature regularly.
Results
NER and AED
We annotated the anthrax-related scientific literature abstracts through PubTator and obtained PBC. PBC contains eight entity categories, 7937 entities and 3715 NCBI_IDs. Then we amplified and corrected PBC to get AED, which includes 29 entity categories, 13 882 entities and 4822 NCBI_IDs. Part content of AED is shown in Table 1. The comparison result between AED and PBC is shown in Figure 4. Figure 5 clearly shows the difference in the number and category of AED and PBC entities. Our mining results are more abundant and specific for anthrax-related biomedical literature.
| ID . | Entity . | Entity_category . | NCBI_ID . |
|---|---|---|---|
| 3 | Anthrax | Disease | MESH:D000881 |
| 5 | Human | Species | 9606 |
| 127 | PA | Toxin | MESH:C030325 |
| 128 | Aluminium hydroxide | Chemical | MESH:D000536 |
| 1577 | IL-1beta | Gene | 16175 |
| 4012 | Threonines | Amino Acid | MESH:D013912 |
| 3356 | Delta724 | DNAMutation | c.del724 |
| 4087 | N719Q | ProteinMutation | p.N719Q |
| 3962 | rs13140055 | SNP | rs13140055 |
| 4299 | SLSV | Vaccine | MESH:D022122 |
| 4680 | o-ATP | Nucleotide | MESH:C017199 |
| 4791 | Caspase-1 | Protein | MESH:D020170 |
| 5253 | rs4690127 | SNP | rs4690127 |
| 5462 | MB49 | CellLine | CVCL:7076 |
| 7493 | Pyrimidine nucleosides | Nucleoside | MESH:D011741 |
| 9158 | Real-time PCR | Technique | MESH:D060888 |
| 9717 | CDC | Agency | MESH:D002487 |
| 10607 | Cell migration | Phenomena | MESH:D002465 |
| 10734 | Eating undercooked meat | Behavior | NULL |
| 11066 | Leu-Leu-OMe | Peptide | MESH:C045139 |
| 11808 | Lung | Anatomy | MESH:D008168 |
| 11938 | Drug therapies | Therapeutics | MESH:D004358 |
| 12029 | capBCADE genes | Genome | 665 |
| 12791 | Half-cooked sheep’s meat | Food | MESH:D008460 |
| 13091 | siRNA | Nucleic Acid | MESH:D034741 |
| 13570 | Intravenous drug user | Person | MESH:D055030 |
| 13656 | Blood pressure | Diagnosis | MESH:D001794 |
| 13835 | Prohormone convertases | Enzyme | MESH:D043484 |
| ID . | Entity . | Entity_category . | NCBI_ID . |
|---|---|---|---|
| 3 | Anthrax | Disease | MESH:D000881 |
| 5 | Human | Species | 9606 |
| 127 | PA | Toxin | MESH:C030325 |
| 128 | Aluminium hydroxide | Chemical | MESH:D000536 |
| 1577 | IL-1beta | Gene | 16175 |
| 4012 | Threonines | Amino Acid | MESH:D013912 |
| 3356 | Delta724 | DNAMutation | c.del724 |
| 4087 | N719Q | ProteinMutation | p.N719Q |
| 3962 | rs13140055 | SNP | rs13140055 |
| 4299 | SLSV | Vaccine | MESH:D022122 |
| 4680 | o-ATP | Nucleotide | MESH:C017199 |
| 4791 | Caspase-1 | Protein | MESH:D020170 |
| 5253 | rs4690127 | SNP | rs4690127 |
| 5462 | MB49 | CellLine | CVCL:7076 |
| 7493 | Pyrimidine nucleosides | Nucleoside | MESH:D011741 |
| 9158 | Real-time PCR | Technique | MESH:D060888 |
| 9717 | CDC | Agency | MESH:D002487 |
| 10607 | Cell migration | Phenomena | MESH:D002465 |
| 10734 | Eating undercooked meat | Behavior | NULL |
| 11066 | Leu-Leu-OMe | Peptide | MESH:C045139 |
| 11808 | Lung | Anatomy | MESH:D008168 |
| 11938 | Drug therapies | Therapeutics | MESH:D004358 |
| 12029 | capBCADE genes | Genome | 665 |
| 12791 | Half-cooked sheep’s meat | Food | MESH:D008460 |
| 13091 | siRNA | Nucleic Acid | MESH:D034741 |
| 13570 | Intravenous drug user | Person | MESH:D055030 |
| 13656 | Blood pressure | Diagnosis | MESH:D001794 |
| 13835 | Prohormone convertases | Enzyme | MESH:D043484 |
| ID . | Entity . | Entity_category . | NCBI_ID . |
|---|---|---|---|
| 3 | Anthrax | Disease | MESH:D000881 |
| 5 | Human | Species | 9606 |
| 127 | PA | Toxin | MESH:C030325 |
| 128 | Aluminium hydroxide | Chemical | MESH:D000536 |
| 1577 | IL-1beta | Gene | 16175 |
| 4012 | Threonines | Amino Acid | MESH:D013912 |
| 3356 | Delta724 | DNAMutation | c.del724 |
| 4087 | N719Q | ProteinMutation | p.N719Q |
| 3962 | rs13140055 | SNP | rs13140055 |
| 4299 | SLSV | Vaccine | MESH:D022122 |
| 4680 | o-ATP | Nucleotide | MESH:C017199 |
| 4791 | Caspase-1 | Protein | MESH:D020170 |
| 5253 | rs4690127 | SNP | rs4690127 |
| 5462 | MB49 | CellLine | CVCL:7076 |
| 7493 | Pyrimidine nucleosides | Nucleoside | MESH:D011741 |
| 9158 | Real-time PCR | Technique | MESH:D060888 |
| 9717 | CDC | Agency | MESH:D002487 |
| 10607 | Cell migration | Phenomena | MESH:D002465 |
| 10734 | Eating undercooked meat | Behavior | NULL |
| 11066 | Leu-Leu-OMe | Peptide | MESH:C045139 |
| 11808 | Lung | Anatomy | MESH:D008168 |
| 11938 | Drug therapies | Therapeutics | MESH:D004358 |
| 12029 | capBCADE genes | Genome | 665 |
| 12791 | Half-cooked sheep’s meat | Food | MESH:D008460 |
| 13091 | siRNA | Nucleic Acid | MESH:D034741 |
| 13570 | Intravenous drug user | Person | MESH:D055030 |
| 13656 | Blood pressure | Diagnosis | MESH:D001794 |
| 13835 | Prohormone convertases | Enzyme | MESH:D043484 |
| ID . | Entity . | Entity_category . | NCBI_ID . |
|---|---|---|---|
| 3 | Anthrax | Disease | MESH:D000881 |
| 5 | Human | Species | 9606 |
| 127 | PA | Toxin | MESH:C030325 |
| 128 | Aluminium hydroxide | Chemical | MESH:D000536 |
| 1577 | IL-1beta | Gene | 16175 |
| 4012 | Threonines | Amino Acid | MESH:D013912 |
| 3356 | Delta724 | DNAMutation | c.del724 |
| 4087 | N719Q | ProteinMutation | p.N719Q |
| 3962 | rs13140055 | SNP | rs13140055 |
| 4299 | SLSV | Vaccine | MESH:D022122 |
| 4680 | o-ATP | Nucleotide | MESH:C017199 |
| 4791 | Caspase-1 | Protein | MESH:D020170 |
| 5253 | rs4690127 | SNP | rs4690127 |
| 5462 | MB49 | CellLine | CVCL:7076 |
| 7493 | Pyrimidine nucleosides | Nucleoside | MESH:D011741 |
| 9158 | Real-time PCR | Technique | MESH:D060888 |
| 9717 | CDC | Agency | MESH:D002487 |
| 10607 | Cell migration | Phenomena | MESH:D002465 |
| 10734 | Eating undercooked meat | Behavior | NULL |
| 11066 | Leu-Leu-OMe | Peptide | MESH:C045139 |
| 11808 | Lung | Anatomy | MESH:D008168 |
| 11938 | Drug therapies | Therapeutics | MESH:D004358 |
| 12029 | capBCADE genes | Genome | 665 |
| 12791 | Half-cooked sheep’s meat | Food | MESH:D008460 |
| 13091 | siRNA | Nucleic Acid | MESH:D034741 |
| 13570 | Intravenous drug user | Person | MESH:D055030 |
| 13656 | Blood pressure | Diagnosis | MESH:D001794 |
| 13835 | Prohormone convertases | Enzyme | MESH:D043484 |
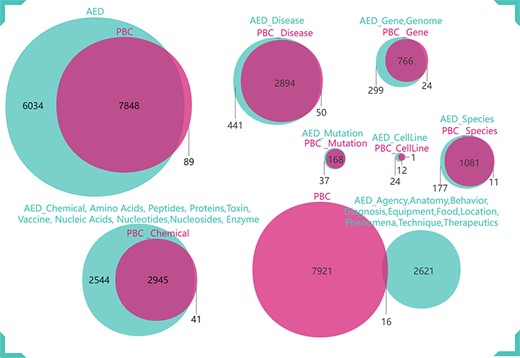
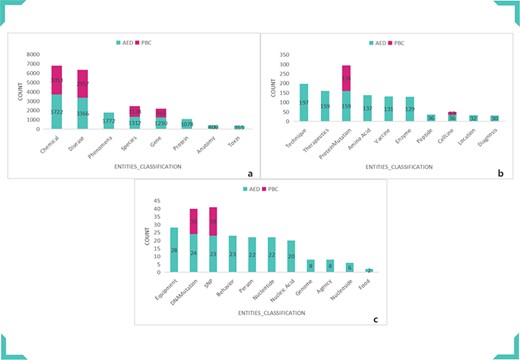
Comparison of AED and PBC entity categories and numbers. (a) Comparison of Chemical, Disease, Phenomena, Species, Gene, Protein, Anatomy and Toxin. (b) Comparison of Technique, Therapeutics, ProteinMutation, Amino Acid, Vaccine, Enzyme, Peptide, CellLine, Location and Diagnosis. (c) Comparison of Equipment, DNAMutation, SNP, Behavior, Person, Nucleotide, Nucleic Acid, Genome, Agency, Nucleoside and Food.
RE and AO
We obtained 7755 triples by RE task combined with manual verification, part of the triples are shown in Table 2. Then we built AO based on the triples combined with AED, and part data of AO is shown in Table 3.
| ID . | Entity1 . | Relation . | Entity2 . | Resource . |
|---|---|---|---|---|
| 1 | Human | Infection by | B. anthracis | 725425|t|Human infection by … |
| 58 | B. anthracis | Is sensitive to | Quinolones | 11770263|B. anthracis is … |
| 1453 | NO production | Was induced by | PGA | 26350415|NO production was … |
| 2031 | Expression of uPA | Is highly correlated to | Tumor invasion | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Against | B. anthracis | 12461034|a|This study aimed to … |
| 4782 | EA1 | Persistently exists in | Spore preparations | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Shows enhanced expression in | Certain tumor endothelia | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Release | Tumor necrosis factor alpha | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Relation . | Entity2 . | Resource . |
|---|---|---|---|---|
| 1 | Human | Infection by | B. anthracis | 725425|t|Human infection by … |
| 58 | B. anthracis | Is sensitive to | Quinolones | 11770263|B. anthracis is … |
| 1453 | NO production | Was induced by | PGA | 26350415|NO production was … |
| 2031 | Expression of uPA | Is highly correlated to | Tumor invasion | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Against | B. anthracis | 12461034|a|This study aimed to … |
| 4782 | EA1 | Persistently exists in | Spore preparations | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Shows enhanced expression in | Certain tumor endothelia | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Release | Tumor necrosis factor alpha | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Relation . | Entity2 . | Resource . |
|---|---|---|---|---|
| 1 | Human | Infection by | B. anthracis | 725425|t|Human infection by … |
| 58 | B. anthracis | Is sensitive to | Quinolones | 11770263|B. anthracis is … |
| 1453 | NO production | Was induced by | PGA | 26350415|NO production was … |
| 2031 | Expression of uPA | Is highly correlated to | Tumor invasion | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Against | B. anthracis | 12461034|a|This study aimed to … |
| 4782 | EA1 | Persistently exists in | Spore preparations | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Shows enhanced expression in | Certain tumor endothelia | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Release | Tumor necrosis factor alpha | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Relation . | Entity2 . | Resource . |
|---|---|---|---|---|
| 1 | Human | Infection by | B. anthracis | 725425|t|Human infection by … |
| 58 | B. anthracis | Is sensitive to | Quinolones | 11770263|B. anthracis is … |
| 1453 | NO production | Was induced by | PGA | 26350415|NO production was … |
| 2031 | Expression of uPA | Is highly correlated to | Tumor invasion | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Against | B. anthracis | 12461034|a|This study aimed to … |
| 4782 | EA1 | Persistently exists in | Spore preparations | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Shows enhanced expression in | Certain tumor endothelia | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Release | Tumor necrosis factor alpha | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Entity_Category . | Entity_id . | Relation . | Resource . |
|---|---|---|---|---|---|
| 1 | Human | Species | 9606 | Infection by | 725425|t|Human infection by … |
| 58 | B. anthracis | Species | 1392 | Is sensitive to | 11770263|B. anthracis is … |
| 1453 | NO production | Phenomena | null | Was induced by | 26350415|NO production was … |
| 2031 | expression of uPA | Phenomena | MESH:D015870 | Is highly correlated to | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Chemical | MESH:C033383 | Against | 12461034|a|This study aimed to … |
| 4782 | EA1 | Gene | 3736 | Persistently exists in | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Gene | 84168 | Shows enhanced expression in | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Anatomy | MESH:D008264 | Release | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Entity_Category . | Entity_id . | Relation . | Resource . |
|---|---|---|---|---|---|
| 1 | Human | Species | 9606 | Infection by | 725425|t|Human infection by … |
| 58 | B. anthracis | Species | 1392 | Is sensitive to | 11770263|B. anthracis is … |
| 1453 | NO production | Phenomena | null | Was induced by | 26350415|NO production was … |
| 2031 | expression of uPA | Phenomena | MESH:D015870 | Is highly correlated to | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Chemical | MESH:C033383 | Against | 12461034|a|This study aimed to … |
| 4782 | EA1 | Gene | 3736 | Persistently exists in | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Gene | 84168 | Shows enhanced expression in | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Anatomy | MESH:D008264 | Release | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Entity_Category . | Entity_id . | Relation . | Resource . |
|---|---|---|---|---|---|
| 1 | Human | Species | 9606 | Infection by | 725425|t|Human infection by … |
| 58 | B. anthracis | Species | 1392 | Is sensitive to | 11770263|B. anthracis is … |
| 1453 | NO production | Phenomena | null | Was induced by | 26350415|NO production was … |
| 2031 | expression of uPA | Phenomena | MESH:D015870 | Is highly correlated to | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Chemical | MESH:C033383 | Against | 12461034|a|This study aimed to … |
| 4782 | EA1 | Gene | 3736 | Persistently exists in | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Gene | 84168 | Shows enhanced expression in | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Anatomy | MESH:D008264 | Release | 15102824|t|Macrophages release … |
| ID . | Entity1 . | Entity_Category . | Entity_id . | Relation . | Resource . |
|---|---|---|---|---|---|
| 1 | Human | Species | 9606 | Infection by | 725425|t|Human infection by … |
| 58 | B. anthracis | Species | 1392 | Is sensitive to | 11770263|B. anthracis is … |
| 1453 | NO production | Phenomena | null | Was induced by | 26350415|NO production was … |
| 2031 | expression of uPA | Phenomena | MESH:D015870 | Is highly correlated to | 11278833|uPAR and uPA are … |
| 3300 | Rokitamycin | Chemical | MESH:C033383 | Against | 12461034|a|This study aimed to … |
| 4782 | EA1 | Gene | 3736 | Persistently exists in | 19915677|EA1 has been well known … |
| 5660 | TEM8 | Gene | 84168 | Shows enhanced expression in | 22912819|TEM8 shows enhanced … |
| 7268 | Macrophages | Anatomy | MESH:D008264 | Release | 15102824|t|Macrophages release … |
AnthraxKG
AnthraxKG contains 6638 nodes of 30 categories, 6492 edges, 32 898 properties and 7055 triples. The full view of AnthraxKG is shown in Figure 6.
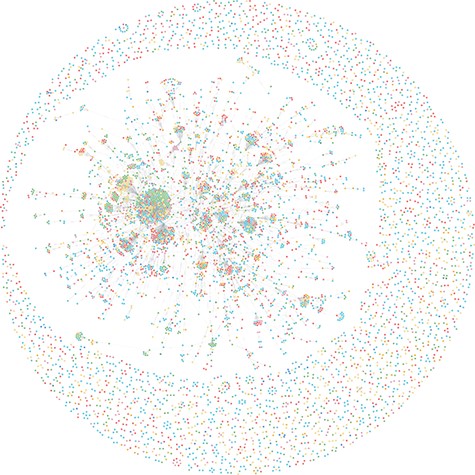
We use the mined anthrax-related entities as the nodes of AnthraxKG. Enter ‘MATCH(Node_Disease: Disease) RETURN Node_Disease’ at the command line of Neo4j Browser to display all the Disease nodes and export the result of the Disease nodes query to a .CSV format file, as shown in Table 4, our entities contain both Name and NCBI_ID properties.
| Node_Disease . |
|---|
| {NCBI_ID:MESH:D003643,Name:lethal toxin-induced death} |
| {NCBI_ID:MESH:D014097,Name:pseudo-anodontia} |
| {NCBI_ID:MESH:D006470,Name:hemorrhagic meningoencephalitis} |
| {NCBI_ID:MESH:D008480,Name:mediastinitis} |
| {NCBI_ID:MESH:D010335,Name:pathologic process} |
| {NCBI_ID:MESH:D017444,Name:skin lesions of human orf} |
| Node_Disease . |
|---|
| {NCBI_ID:MESH:D003643,Name:lethal toxin-induced death} |
| {NCBI_ID:MESH:D014097,Name:pseudo-anodontia} |
| {NCBI_ID:MESH:D006470,Name:hemorrhagic meningoencephalitis} |
| {NCBI_ID:MESH:D008480,Name:mediastinitis} |
| {NCBI_ID:MESH:D010335,Name:pathologic process} |
| {NCBI_ID:MESH:D017444,Name:skin lesions of human orf} |
| Node_Disease . |
|---|
| {NCBI_ID:MESH:D003643,Name:lethal toxin-induced death} |
| {NCBI_ID:MESH:D014097,Name:pseudo-anodontia} |
| {NCBI_ID:MESH:D006470,Name:hemorrhagic meningoencephalitis} |
| {NCBI_ID:MESH:D008480,Name:mediastinitis} |
| {NCBI_ID:MESH:D010335,Name:pathologic process} |
| {NCBI_ID:MESH:D017444,Name:skin lesions of human orf} |
| Node_Disease . |
|---|
| {NCBI_ID:MESH:D003643,Name:lethal toxin-induced death} |
| {NCBI_ID:MESH:D014097,Name:pseudo-anodontia} |
| {NCBI_ID:MESH:D006470,Name:hemorrhagic meningoencephalitis} |
| {NCBI_ID:MESH:D008480,Name:mediastinitis} |
| {NCBI_ID:MESH:D010335,Name:pathologic process} |
| {NCBI_ID:MESH:D017444,Name:skin lesions of human orf} |
We use the extracted relations as the edges of AnthraxKG. Enter in the command line of Neo4j Browser: ‘MATCH Relation=()-->() RETURN Relation’ and export the result of the Relation query to a .CSV format file. It can be seen that the properties of the edges include the properties of the corresponding nodes but also contains the source corresponding to the triples: PMID + Sentence, as shown in Table 5.
| Relation . |
|---|
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:632,Name:Yersinia pestis}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis},{NCBI_ID:MESH:D000881,Name:anthrax}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:1392,Name:Bacillus anthracis}] |
| [{NCBI_ID:MESH:D022122,Name:FDA approved vaccine},{Sentence:32393506|The only currently U.S. FDA approved vaccine to prevent anthrax in humans is Anthrax Vaccine Adsorbed (AVA),},{NCBI_ID:MESH:C493276,Name:Anthrax Vaccine Adsorbed (AVA)}] |
| Relation . |
|---|
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:632,Name:Yersinia pestis}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis},{NCBI_ID:MESH:D000881,Name:anthrax}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:1392,Name:Bacillus anthracis}] |
| [{NCBI_ID:MESH:D022122,Name:FDA approved vaccine},{Sentence:32393506|The only currently U.S. FDA approved vaccine to prevent anthrax in humans is Anthrax Vaccine Adsorbed (AVA),},{NCBI_ID:MESH:C493276,Name:Anthrax Vaccine Adsorbed (AVA)}] |
| Relation . |
|---|
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:632,Name:Yersinia pestis}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis},{NCBI_ID:MESH:D000881,Name:anthrax}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:1392,Name:Bacillus anthracis}] |
| [{NCBI_ID:MESH:D022122,Name:FDA approved vaccine},{Sentence:32393506|The only currently U.S. FDA approved vaccine to prevent anthrax in humans is Anthrax Vaccine Adsorbed (AVA),},{NCBI_ID:MESH:C493276,Name:Anthrax Vaccine Adsorbed (AVA)}] |
| Relation . |
|---|
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:632,Name:Yersinia pestis}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis},{NCBI_ID:MESH:D000881,Name:anthrax}] |
| [{NCBI_ID:MESH:D014612,Name:Bivalent Anthrax-Plague Vaccine},{Sentence:28694806|t|A Bivalent Anthrax-Plague Vaccine That Can Protect against Two Tier-1 Bioterror Pathogens, Bacillus anthracis and Yersinia pestis.},{NCBI_ID:1392,Name:Bacillus anthracis}] |
| [{NCBI_ID:MESH:D022122,Name:FDA approved vaccine},{Sentence:32393506|The only currently U.S. FDA approved vaccine to prevent anthrax in humans is Anthrax Vaccine Adsorbed (AVA),},{NCBI_ID:MESH:C493276,Name:Anthrax Vaccine Adsorbed (AVA)}] |
Discussion
Discussion for the existing data of AO
We will discuss some typical data examples according to the entity categories and the corresponding association relationship.
Anthrax-related species and anthrax-related diseases
We mined 1312 species entities and 3366 disease entities associated with anthrax. The synonymous expressions of anthrax in the related literature are Bacillus anthracis infection, BA infection, B. anthracis infection, infection with Bacillus anthracis, etc., which contain abbreviations, word variations and word order changes. In addition to common herbivorous poultry and wildlife, such as cattle, sheep, pigs, horses and deer that transmit anthrax, we have found that insects including mosquitoes and flies also appear to facilitate the transmission of B. anthracis. Through AnthraxKG, we found that depending on the route of infection anthrax can cause different complications, such as scab ulcers, nausea, coughing up blood, shock, sepsis, hemorrhagic meningitis and toxemia. Also, anthrax epidemics can have psychological effects on people, such as anxiety, excessive alcohol consumption, fear and post-traumatic stress disorder, which also deserve the attention of researchers. We also found that anthrax is associated with the transmission of other zoonotic diseases, for example, anthrax rabies affects each other’s ability to transmit.
Relation between anthrax-related genes and anthrax-related diseases
We mined the text to 1230 anthrax-related gene entities. By AnthraxKG, we found that genes, such as pagA, lef, cya and capA, are involved in the regulation of B. anthracis virulence. Mutations in ANTXR2, the gene encoding the anthrax toxin receptor, cause juvenile hyaline fibromatosis. ANTXR2 is also weakly associated with ankylosing spondylitis. ANTXR1, the gene encoding the anthrax toxin receptor, can be used as a targeted treatment for diseases such as triple-negative breast cancer, gastric cancer and glioma, and it can be combined with passive immunotherapy with CAR T-Cell Therapy for cancer treatment, which is one of the research focuses in the exploration of cancer therapeutic approaches.
Prevention and treatment of anthrax and anthrax-related diseases
We mined 3722 chemical entities, and with AnthraxKG, we found that 106 antibiotics are available for the treatment of anthrax infections by different routes. Bacillus anthracis is sensitive to quinolones, clindamycin, tetracyclines and penicillin. Cutaneous anthrax can be treated with ciprofloxacin an ampicillin-sulbactam drugs. Inhalational anthrax can be treated with drugs such as penicillin and clindamycin. It is suggested that ciprofloxacin or doxycycline can be used as post-exposure prophylaxis for inhalational anthrax. Gastrointestinal anthrax can be treated with penicillin G drugs such as indomethacin or cyclo-oxygenase inhibitors, and neurokinin 12 receptor antagonists can significantly reduce vascular leakage associated with anthrax edema toxin (ET). Levofloxacin, daptomycin, gatifloxacin and dalbavancin emerged as new drugs for treating anthrax. Unexpectedly, we observed that green tea acts as a powerful inhibitor of anthrax lethal factor (LF). As demonstrated by AnthraxKG, the human anthrax vaccines currently in use include Sterne’s strain 34F2 anthrax vaccine, Russian anthrax vaccines, British anthrax vaccines and anthrax vaccine adsorbed (AVA), of which AVA is the only human anthrax vaccine licensed by the FDA in the USA.
Relation between anthrax and chemicals amplified from PBC
We amplified and corrected PBC_chemicals to obtain the entity categories of amino acids, peptides, proteins, enzymes, nucleic acids, nucleotides, nucleosides and toxins. As AnthraxKG showed, Asp187 and Phe190 residues in LF are necessary for the active expression of anthrax lethal toxin. The anthrax toxin secreted by B. anthracis is composed of protective antigen (PA), LF and edema factor. The germination-specific lyases SleB, CwlJ1 and CwlJ2 all contribute to the germination and virulence of B. anthracis spores. The related descriptions of these expanded compounds are most concerned by the medical community for the impact indicators of anthracnose and its related diseases.
Anthrax-related phenomena
We mined 1772 anthrax-related phenomenal entities, including biological phenomena, genetic phenomena, cellular physiological phenomena, signaling pathways and other phenomena and process categories. For example, inactivation of the Pta-AckA pathway impairs B. anthracis adaptation during spillover metabolism. microRNA-493 suppresses hepatocarcinogenesis by downregulating ANTXR1 and R-Spondin 2. Through the relations of phenomena and other entities, the pathogenesis and mechanism of action of anthrax can be presented and help researchers understand the pathogenesis and infection pattern of anthrax, and the phenomena entities serve as an explanatory note to other entities.
Anthrax-related diagnosis, technique and equipment
We mined 197 technologies and 28 devices, which can be used to detect, identify, culture and inactivate B. anthracis spores, for example, electrical graphene aptasensor for sensitive detection of anthrax toxin, and we found real-time loop-mediated isothermal amplification assay can be used for rapid detection of anthrax spores in soil and talcum powder.
Anthrax susceptible location and person category
We have found that anthrax is widespread in the less developed regions of Africa and Central Asia. Drummers, sheep shearers, butchers and others are engaged in breeding or slaughtering livestock and who come into contact with animal products are most susceptible to anthrax infection. We also found that postal workers and members of the US Congress have been infected with anthrax on several occasions in connection with the anthrax attacks that occurred in the USA in 2001. In addition, intravenous drug users can also contract anthrax, such as the anthrax outbreak among drug users in Scotland from December 2009 to December 2010.
Limitations of text mining tools and supplement of manual curation
During the text mining process, we found that some limitations of the mining tools would lead to inaccurate data and that we as authors lacked relevant knowledge in the field. To ensure the data quality of AO and AnthraxKG, we sought help from experts in the field. Professor Weiguang Zhou and his team from the College of Veterinary Medicine, Inner Mongolia Agricultural University, whose main research interests are diagnostic molecular biology of animal pathogens and molecular immunology, undertook the manual curation of the data.
In the NER task, we found that some of the entities extracted by PubTator are inaccurate. For example, ET, which refers to a toxin of B. anthracis, was identified by PubTator as a disease concept ‘edema Disease MESH: D004487’. Another example is that PA also refers to an anthrax toxin, while PubTator did not annotate this entity, and these data need to be checked and corrected. The entity categories also need to be augmented to improve the coverage of the AED. Accomplishing these tasks requires a great deal of expertise, so the experts undertook the curation of the AED.
In the RE task, there are also inaccuracies in the triples extracted by OpenIE. For example, the sentence ‘19654018. anthracis is the etiologic agent of anthrax.’ was processed by OpenIE to obtain four triples, as shown in Figure 7. After cleaning and manual curation by domain experts, we got the triple ‘|-{“subject”: “B. anthracis”, “relation”: “is etiologic agent of”, “object”: “anthrax”}’. For another example, the sentence ‘10597826|Ulnar nerve lesion due to cutaneous anthrax.’ did not get a triple after OpenIE processing. But the sentence did have an association relationship, and after manual curation by experts, we got the triple ‘|-{“subject”: “Ulnar nerve lesion”, “relation”: “due to”, “object”: “cutaneous anthrax”}’.

Conclusions
This study processed anthrax-related scientific literature through text mining tools. Our results show that the work of NER and RE can effectively extract the lots of knowledge hidden in the literature in the biomedical field. Through AO and AnthraxKG, we have made a deeper understanding of anthrax-related diseases, genes, species, chemicals and other related biomedical concepts and interrelationships. We built AnthraxKP and successfully managed and displayed massive amounts of structured data related to anthrax.
In the future research, we will make a continuous effort to update and expand the AO, propose the NER and RE models for anthrax and apply the models to other zoonotic scientific literature mining efforts; furthermore, we intend to incorporate an intelligent question answering algorithm to build an open-source knowledge portal for zoonosis (ZoonosisKP).
Acknowledgements
We would like to thank Professor Weiguang Zhou and his team from Inner Mongolia Agricultural University for their help with the manual curation of the data. Thanks also to Lecturer Zhihong Yang from Inner Mongolia Agricultural University and graduate student Yi Jing from the University of New South Wales for revising this paper for submission in English.
Funding
Inner Mongolia Autonomous Region Natural Science Foundation (2019MS03014); Inner Mongolia Autonomous Region Science and Technology Major Special Projects (2019ZD016, 2020ZD0007, 2021ZD0005); Inner Mongolia Autonomous Region 2021 Graduate Research Innovation Project.
Conflict of interest
None declared.
Author contributions
J.G. designed and supervised the research work. B.F. conducted the text mining work and the development of the knowledge portal.



