-
PDF
- Split View
-
Views
-
Cite
Cite
Bowen Li, Shanshan Yang, Jinjin Long, Xue Chen, Qianyue Zhang, Lin Ning, Bifang He, Heng Chen, Jian Huang, AGODB: a comprehensive domain annotation database of argonaute proteins, Database, Volume 2022, 2022, baac078, https://doi.org/10.1093/database/baac078
Close - Share Icon Share
Abstract
Argonaute (Ago) proteins are widely expressed in almost all organisms. Eukaryotic Ago (eAgo) proteins bind small RNA guides forming RNA-induced silencing complex that silence gene expression, and prokaryotic Ago (pAgo) proteins defend against invading nucleic acids via binding small RNAs or DNAs. pAgo proteins have shown great potential as a candidate ‘scissors’ for gene editing. Protein domains are fundamental units of protein structure, function and evolution; however, the domains of Ago proteins are not well annotated/curated currently. Therefore, full functional domain annotation of Ago proteins is urgently needed for researchers to understand the function and mechanism of Ago proteins. Herein, we constructed the first comprehensive domain annotation database of Ago proteins (AGODB). The database curates detailed information of 1902 Ago proteins, including 1095 eAgos and 807 pAgos. Especially for long pAgo proteins, all six domains are annotated and curated. Gene Ontology (GO) enrichment analysis revealed that Ago genes in different species were enriched in the following GO terms: biological processes (BPs), molecular function and cellular compartment. GO enrichment analysis results were integrated into AGODB, which provided insights into the BP that Ago genes may participate in. AGODB also allows users to search the database with a variety of options and download the search results. We believe that the AGODB will be a useful resource for understanding the function and domain components of Ago proteins. This database is expected to cater to the needs of scientific community dedicated to the research of Ago proteins.
Introduction
Historically, the argonaute (Ago) protein family was discovered in a plant mutagenesis screen (1). Eukaryotic Ago (eAgo) proteins are grouped into four subfamilies: Ago-like, PIWI-like, WAGO and Trypanosoma Ago subfamily (2). We focused on the Ago-like subfamily in this study. eAgo proteins, as the core component of the RNA-induced silencing complex, have become a key player in RNA interference (RNAi) pathways (3). The eAgo proteins mainly form binary complexes with their guide RNAs and recognize complementary target mRNAs for subsequent site-specific cleavage or silencing, which then leads to the degradation and inhibition of protein translation (4). However, due to lack of RNAi pathways in archaea and bacteria, the physiological function of prokaryotic Ago (pAgo) proteins has remained elusive for a long time (5). In recent years, studies have gradually proved that pAgo proteins can bind small RNAs or DNAs to defend against foreign invasive genomes and showed the potential as candidate ‘scissors’ for gene editing (6–9).
eAgo proteins consist of four domains: N-terminal (N), P-element-induced wimpy testis (PIWI)–Ago–Zwille (PAZ), Middle (MID) and PIWI, along with two domain linkers: Linker 1 (L1) and Linker 2 (L2) (10). Each domain is involved in different steps of the protein’s enzyme activity. The N-domain plays an important role in loading and unwinding the small RNA duplex, while the PAZ and MID domains provide binding pockets to anchor the 3ʹ and 5ʹ ends of the microRNA, respectively (11). The PIWI domain is structurally and functionally similar to RNase-H and contributes to endonuclease activity for the target strand (12). However, the endonuclease activity is not a property of all Ago proteins (13, 14). The L1 links the N and PAZ domains, and L2 links the PAZ and MID domains (15). Protein domains are critical in classifying proteins, understanding their biological functions, annotating their evolutionary mechanisms and protein design (16). They are considered to be homologous portions of sequences encoded in different gene contexts, kept intact by evolution at the sequence level (17). Therefore, comprehensive domain annotation of Ago proteins will help researchers to understand the function and mechanism of Ago proteins.
According to the domain architecture, pAgo proteins can be divided into long pAgo proteins, short pAgo proteins and PIWI-RE proteins [PIWI with conserved R (Arg) and E (Glu) residues] (5). The overall domain information of most long pAgo proteins is essentially similar to eAgo proteins, retaining all four domains and two linkers, whereas short pAgo proteins only contain the MID and PIWI domains (18). PIWI-RE proteins with unknown functions appear in several major bacterial lineages, and their domain constituent is similar to short pAgo proteins with two conserved MID and PIWI domains (19). But unlike short pAgo proteins, PIWI-RE proteins have two conserved residues: arginine (R) and glutamate (E) in PIWI and MID domains, which are essential for nucleic acid-binding (4, 19). Currently, although the function and mechanism of eAgo and several long pAgo proteins have been studied in detail, little is known about those of short pAgo and PIWI-RE proteins (20–23).
Protein domains are fundamental units of protein structure, function and evolution (24). At the sequence level, domains are homologous segments in evolution, and at the structural level, domains are units that can fold and work independently (16). Proteins can usually be decomposed into domains according to their similar sequence or structural characteristics. The traditional method of domain annotation is to analyze the structure of Ago proteins by nuclear magnetic resonance, X crystal diffraction and cryo-electron microscopy three-dimensional reconstruction techniques and then divide the protein domains artificially. Using these methods for domain annotation is time-consuming and labor-intensive. Fortunately, Jiang et al. (25) proposed a computational method, called AGONOTES, for comprehensive domain annotation of Ago proteins. However, there is no specific database available for storing domain annotation data of Ago proteins currently. Existing public protein databases, such as the Universal Protein Resource (UniProt) (26), protein families database (Pfam) (27), simple modular architecture research tool (SMART) (28) and Conserved Domain Database (CDD) (29), enable the domain annotation of various proteins, whereas the domains of eAgo proteins have not been well annotated/curated, and only the relatively conservative PIWI and PAZ domains have been annotated and curated. Since the domains of pAgo proteins are not as conserved as that of eAgo proteins, the domain annotation of pAgo protein is more incomplete. Therefore, a comprehensive domain annotation database for Ago proteins is desperately needed.
Herein, we constructed the first comprehensive domain annotation database of Ago proteins (AGODB). The online resource not only provides complete domain annotation but also allows users to search the database with a variety of options and download the search results. In addition, an Ago protein prediction tool named AGOPredict (30) has also been integrated into AGODB to help users to discover novel Ago proteins. Users can access our database at http://i.uestc.edu.cn/agodb/ freely. The database will be of great significance for promoting the research of Ago proteins and related fields.
Materials and methods
Data source
We first searched UniProt with ‘argonaute protein’ as the keyword. We collected all Ago proteins that were manually reviewed and belong to Ago subfamily (namely Ago-like subfamily). Meanwhile, we manually curated the Ago proteins from the published literature. Ago proteins collected from UniProt and published literature with three-dimensional structures resolved experimentally were considered as experimentally validated Ago proteins. Then, we collected the general information corresponding to each protein from UniProt and National Center for Biotechnology Information (NCBI) (31) and the domain information corresponding to each protein from published literature. For those Ago proteins with no domain information in the literature, we used the domain annotation tool of Ago proteins named AGONOTES to make comprehensive domain prediction and collected the results into our database. To find more candidate Ago proteins, we then put the sequences of the experimentally verified Ago proteins into the blastp tool in BLAST 2.12.0+ for similarity search with default parameters (32). Based on the similarity search results, the protein was identified as a potential Ago protein when the identity was greater than 90%, the e-value was less than 1e − 5 and the coverage identity was greater than 85%. After confirming that the protein was not in the Ago protein data set already collected, we curated it into AGODB. To provide a non-redundant Ago protein data set, we utilized the CD-HIT tool to remove redundant sequences with a sequence identity cutoff of 80% (33). Users can download this data set at http://i.uestc.edu.cn/agodb/download.html.
GO and KEGG pathway analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and Gene Ontology (GO) functional annotation can preliminarily analyze the biological process (BP) or signal pathway that genes may participate in. We used KOBAS-intelligence (KOBAS-i) (34) to perform gene annotation and pathway enrichment analysis. Significant GO terms and pathways were identified after multiple testing adjustments using the false discovery rate (FDR) method; FDR-corrected P-value < 0.05 indicated a statistically significant difference.
Database implementation
The front end of the AGODB website was built with the languages HTML and PHP. MySQL, which is one of the most popular relational database management systems, was used for data storage and processing in the backend. The architecture of AGODB is shown in Figure 1. Browsing and searching panels were developed for visiting and retrieving data from the database efficiently. AGODB has been tested in three popular web browsers, including the Google Chrome, Internet Explorer and Mozilla Firefox browsers.
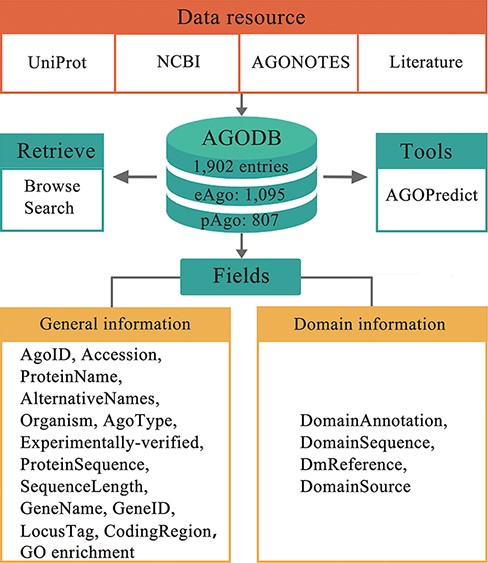
Architecture of AGODB. AGODB consists of 1902 entries, including 1095 eAgo proteins and 807 pAgo proteins. The relevant information of each entry in AGODB was collected from UniProt, NCBI, AGONOTES and published articles. The development of AGODB includes the design of fields, the development of the retrieval system and the integration of tool(s).
Tools integration
AGOPredict, which is a support vector machine-based predictor specifically for identifying Ago proteins based on sequence information (30), was integrated into AGODB. This bioinformatics tool will help users to discover more potential Ago proteins, and users can use it for Ago protein prediction by clicking ‘AGOPREDICT’ at the top of AGODB. In addition, we also provide links to AGONOTES and AGO3D on the homepage of AGODB to facilitate users to study Ago proteins. AGONOTES (25) is an online service dedicated to annotating the comprehensive domains of Ago proteins, and AGO3D (35) is an online service tailored for constructing three-dimensional structures of Ago proteins.
Results
Data content and summary
In AGODB, the collected information was divided into two modules: general information and domain annotation.
General information
Information on Ago proteins was manually obtained from UniProt or NCBI and stored as general information. Each entry included the following information: (i) AgoID—the unique number of the Ago protein in the database; (ii) Accession—the accession number of the Ago protein in NCBI, with a link to the NCBI database. If the accession number of the Ago protein cannot be linked to NCBI, then linked to UniProt; (iii) ProteinName—the name of the Ago protein; (iv) AlternativeNames—synonyms of the Ago protein name; (v) Organism—the name of organism that is the source of the protein sequence; (vi) AgoType—eAgo or pAgo; (vii) Experimentally verified—whether it has been experimentally verified; (viii) ProteinSequence—the amino acid sequence of the Ago protein named after the accession number, with a link to fasta format sequence in the NCBI protein database. If the accession number of the Ago protein cannot be linked to NCBI, then linked to UniProt. (ix) SequenceLength—the number of amino acids; (x) GeneName—the name of the gene that codes for the protein sequence, with its synonyms; (xi) GeneID—a unique number for each gene, with a link to the NCBI gene database; (xii) LocusTag—the identifier in the genome; (xiii) CodingRegion—coding region; (xiv) GO enrichment—GO terms which provide insights into the BP that Ago genes may participate in.
Domain information
On the basis of general information, the important domain information of Ago proteins was collected. Domain information from the literature or annotated by AGONOTES was manually curated. This information consisted of four fields, including (i) DomainAnnotation: the domain annotation information of the Ago protein displayed in the form of a figure. (ii) DomainSequence: a download button to retrieve the complete sequence of each domain. For parts of Ago proteins, due to the scope of each domain from the literature is not clear, the domain sequence is not available. (iii) DmReference: reference for domain annotation. (iv) DomainSource: the source of the domain annotation information (AGONOTES or literature).
The current release of AGODB holds related information about Ago proteins, with a total of 1902 entries. As shown in Figure 2A, 70 Ago proteins are experimentally validated, and the others are putative Ago proteins that have yet to be further confirmed. As shown in Figure 2B, 1095 Ago proteins are derived from eukaryotes and 807 are derived from prokaryotes. As depicted in Figure 3, 1377 Ago proteins have complete domain annotation information, of which 1366 are annotated by AGONOTES and 11 are from published articles; 512 Ago proteins have incomplete domain information which was provided by Ryazansky et al. (18) and 13 Ago proteins had no domain information.
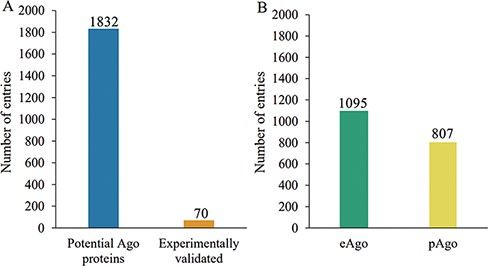
Schematic of Ago protein entries at various categories in the AGODB database. (A) Number of potential and experimentally validated Ago proteins. (B) Number of eAgo and pAgo proteins.
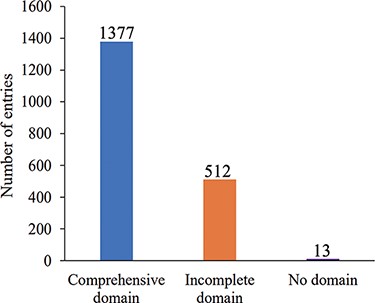
Number distribution of Ago protein entries according to domain annotation information integrity.
Data browsing and searching
A user-friendly browsing interface has been developed. By clicking ‘BROWSE’ at the top of the AGODB website, users first access a brief browsing table that displays fields, including AgoID, Accession, ProteinName, Organism, AgoType, Experimentally verified and links guiding to the ‘Detail’ page (Figure 4A). As shown in Figure 4B, detailed descriptions are provided in the ‘Detail’ page, which can be reached by selecting ‘more’ in the browsing page.
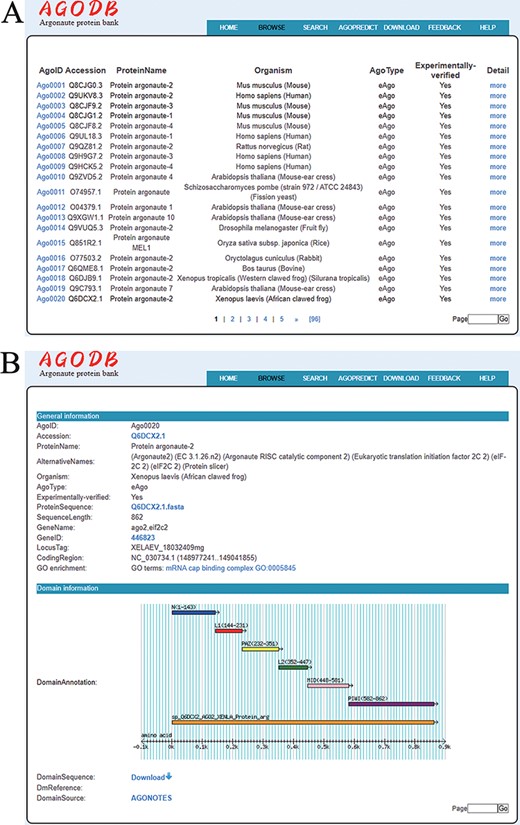
Browsing page of AGODB. (A) A brief browsing table displays fields, including AgoID, Accession, ProteinName, Organism, AgoType, Experimentally verified and links directing to the ‘Detail’ page. (B) The ‘Detail’ page shows detailed descriptions of each Ago protein.
AGODB also provides a search system that allows users to easily retrieve data by clicking ‘SEARCH’ at the top of website or ‘Search System’ on the home page. As shown in Figure 5, users can submit a single query against most fields of the database, such as AgoID, Accession, ProteinName, Organism, AgoType, Experimentally verified, GeneID, etc., and also can submit multiple queries simultaneously with Boolean expressions (e.g. AND and OR). The search results are displayed on the search result page in tabular form, similar to browsing the database. More detailed descriptions are displayed in the ‘Detail’ page, which can be reached by clicking ‘more’. Simultaneously, the search results can be conveniently downloaded in batch in *.xml or *.csv format.
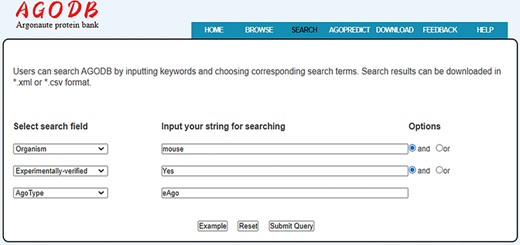
Search interface of AGODB. For example, if users want to retrieve all the experimentally validated eAgo proteins from mouse, they should first select the search field ‘Organism’ from the pull-down list box, input ‘mouse’ into the corresponding blank text form and select the ‘and’ button. Then, users should select the ‘Experimentally-verified’ search field, input ‘Yes’ and select the ‘and’ button. Finally, they should select the ‘AgoType’ field, input ‘eAgo’ and press the ‘Submit Query’ button.
GO and KEGG pathway analysis
In AGODB, we collected a total of 1902 proteins, which are from 1068 species. ‘Gene ID’ can be found in 842 Ago proteins. We performed GO functional annotation and KEGG pathway enrichment analysis using KOBAS-i (34). We kept the results with P-value < 0.05 (FDR corrected) and finally got the results of GO functional annotation for 11 species. Ago genes in species Arabidopsis thaliana, Bos taurus, Danio rerio, Drosophila melanogaster, Gallus gallus, Homo sapiens, Mus musculus, Rattus norvegicus, Sus scrofa and Xenopus tropicalis were enriched in the following GO terms: BPs, molecular function and cellular compartment (Supplementary Tables S1–S11), and the top five subclasses of GO enrichment terms in species Arabidopsis thaliana and Bos taurus are shown in Figure 6. The top five subclasses of GO enrichment terms in the other nine species are depicted in Supplementary Figures 1–5. We integrated the results of GO function annotation into AGODB. However, no results were obtained from the KEGG pathway enrichment analysis.
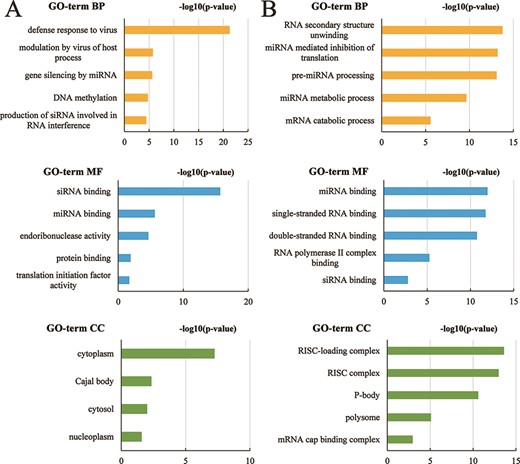
The top five subclasses of GO enrichment terms. (A) Arabidopsis thaliana species. (B) Bos taurus species. BP: biological processes; MF: molecular function; CC: cellular component.
Discussion and conclusion
During the past 20 years, significant progress has been made in understanding the structural and biochemical function of the Ago superfamily (4). In recent years, Ago proteins have received extensive attention for their potential applications in gene editing. Researchers are still looking for Ago proteins that can work at moderate temperatures and are suitable for gene editing. Hence, AGODB not only can provide candidates for developing gene editing tools but also has important significance for further promoting the data mining of Ago proteins. In AGODB, comprehensive domain annotation of Ago proteins is mainly provided, followed by general information of Ago proteins and the BPs that Ago genes may participate in.
We encountered some challenges when collecting domain information of Ago proteins. There were only 11 Ago proteins with comprehensive domain annotation that can be manually curated from the literature, and incomplete domain information of 512 Ago proteins was provided by Ryazansky et al. (18). Therefore, we took advantage of a well-performing domain annotation tool called AGONOTES to predict the domain of the remaining Ago proteins. AGONOTES annotated comprehensive domain of 1366 Ago proteins. However, there are still 13 Ago proteins’ domains that cannot be annotated by AGONOTES.
In conclusion, we provided the first database special for Ago proteins, AGODB, with data retrieval capabilities and browsing, searching and downloading options to facilitate data access. This bioinformatics resource contains detailed information of Ago proteins, including 1095 eAgos and 807 pAgos. AGODB is freely available at http://i.uestc.edu.cn/agodb/. In the future, we will continue to collect the related information of Ago proteins and update AGODB.
Supplementary data
Supplementary data are available at Database Online.
Acknowledgements
The authors are grateful to the anonymous reviewers for their valuable suggestions and comments, which will lead to the improvement of this paper.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers: 61901130, 61901129 and 62071099), the Guizhou Provincial Science and Technology Projects [grant nos. ZK(2022)-general-056, ZK(2022)-general-038, (2020)1Y407 and (2020)1Y345], Health Commission of Guizhou Province (grant no: gzwkj2022-473) and Guizhou University [grant nos. (2018)54, (2018)55 and (2020)5].
Author’s contributions
B.L., S.Y., J.L., X.C., Q.Z. and L.N. developed the web interface of the database. B.L. and S.Y. collected and curated the data. B.L., S.Y. and B.H. wrote the manuscript. B.L., S.Y., B.H., H.C. and J.H. conceived the idea and coordinated the project.
Conflict of interest
There have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
http://i.uestc.edu.cn/agodb/index.html.
References
UniProt, C. (
Author notes
contributed equally to this work.



