-
PDF
- Split View
-
Views
-
Cite
Cite
Xi Zhang, Yining Hu, David Roy Smith, HSDatabase—a database of highly similar duplicate genes from plants, animals, and algae, Database, Volume 2022, 2022, baac086, https://doi.org/10.1093/database/baac086
Close - Share Icon Share
Abstract
Gene duplication is an important evolutionary mechanism capable of providing new genetic material, which in some instances can help organisms adapt to various environmental conditions. Recent studies, for example, have indicated that highly similar duplicate genes (HSDs) are aiding adaptation to extreme conditions via gene dosage. However, for most eukaryotic genomes HSDs remain uncharacterized, partly because they can be hard to identify and categorize efficiently and effectively. Here, we collected and curated HSDs in nuclear genomes from various model animals, land plants and algae and indexed them in an online, open-access sequence repository called HSDatabase. Currently, this database contains 117 864 curated HSDs from 40 distinct genomes; it includes statistics on the total number of HSDs per genome as well as individual HSD copy numbers/lengths and provides sequence alignments of the duplicate gene copies. HSDatabase also allows users to download sequences of gene copies, access genome browsers, and link out to other databases, such as Pfam and Kyoto Encyclopedia of Genes and Genomes. What is more, a built-in Basic Local Alignment Search Tool option is available to conveniently explore potential homologous sequences of interest within and across species. HSDatabase has a user-friendly interface and provides easy access to the source data. It can be used on its own for comparative analyses of gene duplicates or in conjunction with HSDFinder, a newly developed bioinformatics tool for identifying, annotating, categorizing and visualizing HSDs.
Database URL: http://hsdfinder.com/database/
Introduction
Gene duplication is a near-ubiquitous phenomenon throughout the eukaryotic tree of life (1), one that can be advantageous or disadvantageous, depending on the circumstances. For example, under certain conditions, it can be detrimental for an organism to retain highly similar expressed genes (2). Thus, with notable exceptions, it is relatively rare for species to maintain duplicate genes encoding the same functions (3). Nevertheless, it is becoming more apparent that in some situations the generation and maintenance of highly similar duplicate genes (HSDs) is possible, particularly for genes encoding products that are in high demand, such as histones or ribosomal proteins (4). Indeed, there are many examples suggesting that genes involved in stress response, sensory functions, transport and/or metabolism are likely to be fixed as duplicated copies given specific environmental conditions (5).
Recently, Zhang et al. (6) revealed that hundreds of HSDs, involved in diverse cellular processes, are maintained in the psychrophilic Antarctic green alga Chlamydomonas sp. UWO241, which was recently renamed Chlamydomonas priscuii (7). It is believed that these HSDs are aiding its survival via gene dosage (8). Unfortunately, the HSDs from most other eukaryotic genomes, particularly those of algae, remain uncharacterized. This is partly because the experimental methods for identifying HSDs are time-consuming and labor-intensive. Many of the available bioinformatics tools for characterizing homologs are limited by their designs (e.g. they only identify orthologs) or their specificity (e.g. they only identify retrocopies or co-localized duplicates) (9–13). Consequently, we recently developed a web-based tool called HSDFinder that can identify HSDs in eukaryotic genomes with high accuracy and reliability (14). For example, HSDFinder predicted 336 and 265 HSDs in the psychrophilic green algae UWO241 and Chlamydomonas sp. ICE-L (6), respectively, which is consistent with other experimental data (8). By applying HSDFinder to a variety of other species (15), we predicted and cataloged thousands of HSD candidates, which are now curated and documented in a new online repository called HSDatabase. Currently, it houses 117 864 HSDs from 40 eukaryotic species, with a focus on green algae, animals and land plants.
Here, we briefly introduce the general features as well as the procedures and principles for collecting data from HSDatabase. In short, HSDatabase contains information on HSD number, gene copy number and gene copy length. Additionally, the protein functional domains and associated pathways of the HSDs can be retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and InterProScan (16). A built-in Basic Local Alignment Search Tool (BLAST)-search option is also provided, allowing users to conveniently explore potential homologous sequences of interest within and across species. HSDatabase also provides data on a range of other parameters about gene duplicates, such as the number of HSD per Mb, the most commonly conserved domains among HSDs and the functional categories of HSDs. It is our hope to build a comparative analysis framework across species, especially for best-assembled eukaryotic genomes from species living in extreme environments, to better understand the role of gene duplication in adaptive evolution.
Materials and methods
Database collection
HSDs were identified in 40 well-assembled nuclear genomes from diverse model species, including land plants (e.g. Arabidopsis thaliana and Zea mays), algae (e.g. Chlamydomonas reinhardtii and Fragilariopsis cylindrus) and animals (e.g. Drosophila melanogaster, Homo sapiens and Mus musculus) (Figure 1). We focused on model animal and plant genomes because of their high-quality assemblies and annotations. The genome sequences of the selected species are all retrievable from the National Center for Biotechnology Information (NCBI) (17) (Table 1). The HSDs, which are represented by gene copies with nearly identical lengths and similar gene structures, were identified using HSDFinder (14). The identification method is based on all-against-all BLASTP analyses (18) carried out using uniform homology assessment metrics: E-value cut-off ≤1e−10, amino acid pairwise identity ≥90% and amino acid aligned length variance ≤10. Note, the short form of these parameters is denoted as ‘90%_10aa’. Additionally, putative HSDs were expected to have similar structural information, such as matching protein family (Pfam) domains (19), corresponding InterPro annotations (16) and/or nearly identical conserved residues. The InterProScan tool (16), which is an integrated platform for protein signatures, was used to collect the structural information of the HSDs. The all-against-all BLAST and InterProScan results (tab-delimited files) were fed into HSDFinder to generate HSD candidates in an 8-column tab-delimited file (Figure 2A). These candidates were identified by parsing the BLAST all-against-all protein similarity search results with the homology metrics: amino acid pairwise identity and amino acid aligned length variance. To collect and curate the data in HSDatabase, we performed a series of combo thresholds for filtering putatively functional gene copies (described below at Database curation section).
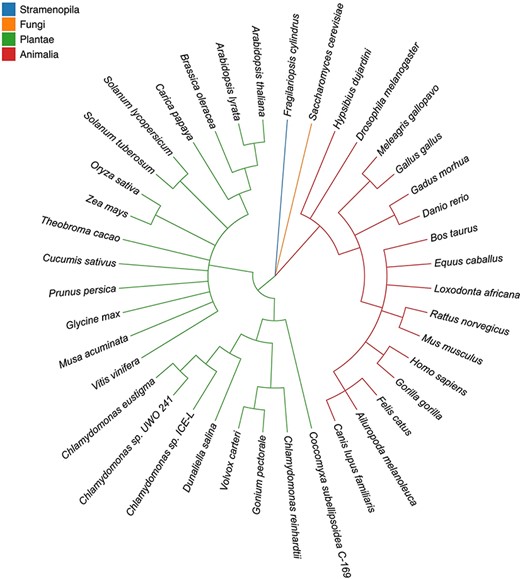
Taxonomic tree of 40 eukaryotic species in four highlighted categories. Stramenopila, Plantae , Fungi and Animalia are in blue, orange, green and red, respectively. The tree topologies were inferred by Taxonomy Common Tree from NCBI (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi).
Summary statistics of the curated HSD groups in the selected genomes from HSDatabase
| Species name (common name) . | Classification . | Curated HSD groups # . | Genome size (Mb) . | No. of considered genes . | Gene copies . | HSDs/genes . | HSDs/Mb . | 2-group HSDsa . | 3-group HSDs . | ≥4-group HSDs . | Genome assembly accession numberb . | Ref . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ailuropoda melanoleuca (giant panda) | Animalia | 2534 | 2371.8 | 22 450 | 8091 | 0.113 | 1.068 | 1570 | 440 | 524 | GCF_002007445.1 | (23) |
| Bos taurus (cattle) | Animalia | 2238 | 2667.85 | 21 036 | 7399 | 0.106 | 0.839 | 1433 | 433 | 372 | GCF_002263795.1 | (24) |
| Canis lupus familiaris (dog) | Animalia | 2372 | 2344.09 | 20 945 | 7314 | 0.113 | 1.012 | 1541 | 434 | 397 | GCF_014441545.1 | (25) |
| Danio rerio (zebrafish) | Animalia | 3799 | 1405.1 | 26 439 | 11 930 | 0.144 | 2.704 | 2263 | 689 | 847 | GCF_000002035.6 | (26) |
| Drosophila melanogaster (fruit fly) | Animalia | 782 | 138.93 | 13 739 | 2230 | 0.057 | 5.629 | 557 | 118 | 107 | GCF_000001215.4 | (27) |
| Equus caballus (horse) | Animalia | 2403 | 2474.93 | 21 126 | 7499 | 0.114 | 0.971 | 1509 | 468 | 426 | GCF_002863925.1 | (28) |
| Felis catus (domestic cat) | Animalia | 2167 | 2493.14 | 20 452 | 6691 | 0.106 | 0.869 | 1409 | 396 | 362 | GCF_018350175.1 | (29) |
| Gadus morhua (Atlantic cod) | Animalia | 3269 | 627.04 | 23 482 | 9757 | 0.139 | 5.213 | 1969 | 633 | 667 | GCF_902167405.1 | (30) |
| Gallus gallus (chicken) | Animalia | 2026 | 1037.17 | 17 797 | 6092 | 0.114 | 1.953 | 1323 | 374 | 329 | GCF_016699485.2 | (31) |
| Gorilla gorilla (western gorilla) | Animalia | 2335 | 3063.36 | 20 632 | 6683 | 0.113 | 0.762 | 1531 | 434 | 370 | GCF_008122165.1 | (32) |
| Homo sapiens (human) | Animalia | 2178 | 2864.14 | 19 531 | 6352 | 0.112 | 0.760 | 1442 | 395 | 341 | GCF_000001405.39 | (33) |
| Hypsibius dujardini (waterbear) | Animalia | 2290 | 182.15 | 20 853 | 5710 | 0.110 | 12.572 | 1618 | 366 | 306 | GCA_002082055.1 | (34) |
| Loxodonta africana (African savanna elephant) | Animalia | 2031 | 3196.74 | 21 094 | 7371 | 0.096 | 0.635 | 1327 | 360 | 344 | GCF_000001905.1 | (35) |
| Meleagris gallopavo (turkey) | Animalia | 1486 | 1058.64 | 17 974 | 3968 | 0.083 | 1.404 | 1061 | 240 | 185 | GCF_000146605.3 | (36) |
| Mus musculus (house mouse) | Animalia | 2402 | 2588.62 | 30 736 | 8855 | 0.108 | 0.928 | 1480 | 459 | 463 | GCF_000001635.27 | (37) |
| Rattus norvegicus (Norway rat) | Animalia | 2434 | 2647.92 | 22 219 | 8757 | 0.110 | 0.919 | 1478 | 471 | 485 | GCF_015227675.2 | (38) |
| Saccharomyces cerevisiae (yeast) | Fungi | 397 | 11.83 | 6002 | 1006 | 0.073 | 33.559 | 331 | 41 | 25 | GCA_003086655.1 | (39) |
| Arabidopsis lyrata | Plantae | 5302 | 202.97 | 29 817 | 16 901 | 0.178 | 26.122 | 3104 | 985 | 1213 | GCF_000004255.2 | (40) |
| Arabidopsis thaliana (thale cress) | Plantae | 4428 | 119.75 | 27 560 | 14 225 | 0.161 | 36.977 | 2630 | 793 | 1005 | GCF_000001735.4 | (41) |
| Brassica oleracea (wild cabbage) | Plantae | 8918 | 529.92 | 44 382 | 30 511 | 0.201 | 16.829 | 4565 | 1841 | 2512 | GCF_000695525.1 | (42) |
| Carica papaya (papaya) | Plantae | 2094 | 360.63 | 18 126 | 6311 | 0.116 | 5.807 | 1299 | 374 | 421 | GCF_000150535.2 | (43) |
| Chlamydomonas eustigma (green alga) | Plantae | 966 | 66.63 | 14 161 | 2199 | 0.068 | 14.498 | 778 | 109 | 79 | GCA_002335675.1 | (20) |
| Chlamydomonas reinhardtii (green alga) | Plantae | 1129 | 111.11 | 19 870 | 3160 | 0.064 | 10.161 | 740 | 187 | 202 | GCF_000002595.2 | (44) |
| Chlamydomonas sp. ICE-L | Plantae | 1540 | 541.86 | 17 731 | 3853 | 0.078 | 2.842 | 1139 | 224 | 177 | GCA_013435795.1 | (21) |
| Chlamydomonas sp. UWO 241 (green alga) | Plantae | 1112 | 211.64 | 16 018 | 3282 | 0.068 | 5.254 | 741 | 139 | 232 | GCA_016618255.1 | (6) |
| Coccomyxa subellipsoidea C-169 (green alga) | Plantae | 360 | 48.83 | 9839 | 1015 | 0.037 | 7.373 | 281 | 43 | 36 | GCA_000258705.1 | (45) |
| Cucumis sativus (cucumber) | Plantae | 2891 | 240.99 | 20 038 | 9558 | 0.144 | 11.996 | 1655 | 532 | 704 | GCF_000004075.3 | (46) |
| Dunaliella salina (green alga) | Plantae | 1589 | 343.7 | 18 740 | 3859 | 0.095 | 4.623 | 1227 | 194 | 168 | GCA_002284615.2 | (47) |
| Fragilariopsis cylindrus (diatom) | Stramenopila | 1129 | 74.76 | 18 111 | 3192 | 0.062 | 15.102 | 766 | 172 | 191 | GCA_001750085.1 | (48) |
| Glycine max (soybean) | Plantae | 11 107 | 995.27 | 47 064 | 38 274 | 0.236 | 11.160 | 6559 | 1295 | 3253 | GCF_000004515.6 | (49) |
| Gonium pectorale (green alga) | Plantae | 1028 | 148.81 | 16 290 | 2669 | 0.063 | 6.908 | 719 | 143 | 166 | GCA_001584585.1 | (50) |
| Musa acuminata (dwarf banana) | Plantae | 5489 | 461.54 | 22 177 | 20 934 | 0.179 | 11.893 | 2633 | 1083 | 1773 | GCF_000313855.2 | (51) |
| Oryza sativa (rice) | Plantae | 4531 | 386.49 | 28 735 | 14 704 | 0.158 | 11.723 | 2641 | 812 | 1078 | GCF_001433935.1 | (52) |
| Prunus persica (peach) | Plantae | 3454 | 220.9 | 23 133 | 11 210 | 0.149 | 15.636 | 2013 | 611 | 830 | GCF_000346465.2 | (53) |
| Solanum lycopersicum (tomato) | Plantae | 4144 | 809.18 | 25 612 | 13 711 | 0.162 | 5.121 | 2345 | 768 | 1031 | GCF_000188115.4 | (54) |
| Solanum tuberosum (potato) | Plantae | 4733 | 768.2 | 28 407 | 15 926 | 0.167 | 6.161 | 2647 | 880 | 1206 | GCF_000226075.1 | (55) |
| Theobroma cacao (cacao) | Plantae | 3074 | 335.44 | 21 517 | 9933 | 0.143 | 9.164 | 1775 | 554 | 745 | GCF_000208745.1 | (56) |
| Vitis vinifera (wine grape) | Plantae | 4039 | 427.21 | 25 830 | 13 613 | 0.156 | 9.454 | 2293 | 722 | 1024 | GCF_000003745.3 | (57) |
| Volvox carteri (green alga) | Plantae | 863 | 137.68 | 14 436 | 2600 | 0.061 | 6.268 | 509 | 152 | 202 | GCA_000143455.1 | (58) |
| Zea mays (Maize) | Plantae | 6801 | 2191.6 | 34 328 | 22 499 | 0.198 | 3.103 | 3910 | 1146 | 1745 | GCF_902167145.1 | (59) |
| Species name (common name) . | Classification . | Curated HSD groups # . | Genome size (Mb) . | No. of considered genes . | Gene copies . | HSDs/genes . | HSDs/Mb . | 2-group HSDsa . | 3-group HSDs . | ≥4-group HSDs . | Genome assembly accession numberb . | Ref . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ailuropoda melanoleuca (giant panda) | Animalia | 2534 | 2371.8 | 22 450 | 8091 | 0.113 | 1.068 | 1570 | 440 | 524 | GCF_002007445.1 | (23) |
| Bos taurus (cattle) | Animalia | 2238 | 2667.85 | 21 036 | 7399 | 0.106 | 0.839 | 1433 | 433 | 372 | GCF_002263795.1 | (24) |
| Canis lupus familiaris (dog) | Animalia | 2372 | 2344.09 | 20 945 | 7314 | 0.113 | 1.012 | 1541 | 434 | 397 | GCF_014441545.1 | (25) |
| Danio rerio (zebrafish) | Animalia | 3799 | 1405.1 | 26 439 | 11 930 | 0.144 | 2.704 | 2263 | 689 | 847 | GCF_000002035.6 | (26) |
| Drosophila melanogaster (fruit fly) | Animalia | 782 | 138.93 | 13 739 | 2230 | 0.057 | 5.629 | 557 | 118 | 107 | GCF_000001215.4 | (27) |
| Equus caballus (horse) | Animalia | 2403 | 2474.93 | 21 126 | 7499 | 0.114 | 0.971 | 1509 | 468 | 426 | GCF_002863925.1 | (28) |
| Felis catus (domestic cat) | Animalia | 2167 | 2493.14 | 20 452 | 6691 | 0.106 | 0.869 | 1409 | 396 | 362 | GCF_018350175.1 | (29) |
| Gadus morhua (Atlantic cod) | Animalia | 3269 | 627.04 | 23 482 | 9757 | 0.139 | 5.213 | 1969 | 633 | 667 | GCF_902167405.1 | (30) |
| Gallus gallus (chicken) | Animalia | 2026 | 1037.17 | 17 797 | 6092 | 0.114 | 1.953 | 1323 | 374 | 329 | GCF_016699485.2 | (31) |
| Gorilla gorilla (western gorilla) | Animalia | 2335 | 3063.36 | 20 632 | 6683 | 0.113 | 0.762 | 1531 | 434 | 370 | GCF_008122165.1 | (32) |
| Homo sapiens (human) | Animalia | 2178 | 2864.14 | 19 531 | 6352 | 0.112 | 0.760 | 1442 | 395 | 341 | GCF_000001405.39 | (33) |
| Hypsibius dujardini (waterbear) | Animalia | 2290 | 182.15 | 20 853 | 5710 | 0.110 | 12.572 | 1618 | 366 | 306 | GCA_002082055.1 | (34) |
| Loxodonta africana (African savanna elephant) | Animalia | 2031 | 3196.74 | 21 094 | 7371 | 0.096 | 0.635 | 1327 | 360 | 344 | GCF_000001905.1 | (35) |
| Meleagris gallopavo (turkey) | Animalia | 1486 | 1058.64 | 17 974 | 3968 | 0.083 | 1.404 | 1061 | 240 | 185 | GCF_000146605.3 | (36) |
| Mus musculus (house mouse) | Animalia | 2402 | 2588.62 | 30 736 | 8855 | 0.108 | 0.928 | 1480 | 459 | 463 | GCF_000001635.27 | (37) |
| Rattus norvegicus (Norway rat) | Animalia | 2434 | 2647.92 | 22 219 | 8757 | 0.110 | 0.919 | 1478 | 471 | 485 | GCF_015227675.2 | (38) |
| Saccharomyces cerevisiae (yeast) | Fungi | 397 | 11.83 | 6002 | 1006 | 0.073 | 33.559 | 331 | 41 | 25 | GCA_003086655.1 | (39) |
| Arabidopsis lyrata | Plantae | 5302 | 202.97 | 29 817 | 16 901 | 0.178 | 26.122 | 3104 | 985 | 1213 | GCF_000004255.2 | (40) |
| Arabidopsis thaliana (thale cress) | Plantae | 4428 | 119.75 | 27 560 | 14 225 | 0.161 | 36.977 | 2630 | 793 | 1005 | GCF_000001735.4 | (41) |
| Brassica oleracea (wild cabbage) | Plantae | 8918 | 529.92 | 44 382 | 30 511 | 0.201 | 16.829 | 4565 | 1841 | 2512 | GCF_000695525.1 | (42) |
| Carica papaya (papaya) | Plantae | 2094 | 360.63 | 18 126 | 6311 | 0.116 | 5.807 | 1299 | 374 | 421 | GCF_000150535.2 | (43) |
| Chlamydomonas eustigma (green alga) | Plantae | 966 | 66.63 | 14 161 | 2199 | 0.068 | 14.498 | 778 | 109 | 79 | GCA_002335675.1 | (20) |
| Chlamydomonas reinhardtii (green alga) | Plantae | 1129 | 111.11 | 19 870 | 3160 | 0.064 | 10.161 | 740 | 187 | 202 | GCF_000002595.2 | (44) |
| Chlamydomonas sp. ICE-L | Plantae | 1540 | 541.86 | 17 731 | 3853 | 0.078 | 2.842 | 1139 | 224 | 177 | GCA_013435795.1 | (21) |
| Chlamydomonas sp. UWO 241 (green alga) | Plantae | 1112 | 211.64 | 16 018 | 3282 | 0.068 | 5.254 | 741 | 139 | 232 | GCA_016618255.1 | (6) |
| Coccomyxa subellipsoidea C-169 (green alga) | Plantae | 360 | 48.83 | 9839 | 1015 | 0.037 | 7.373 | 281 | 43 | 36 | GCA_000258705.1 | (45) |
| Cucumis sativus (cucumber) | Plantae | 2891 | 240.99 | 20 038 | 9558 | 0.144 | 11.996 | 1655 | 532 | 704 | GCF_000004075.3 | (46) |
| Dunaliella salina (green alga) | Plantae | 1589 | 343.7 | 18 740 | 3859 | 0.095 | 4.623 | 1227 | 194 | 168 | GCA_002284615.2 | (47) |
| Fragilariopsis cylindrus (diatom) | Stramenopila | 1129 | 74.76 | 18 111 | 3192 | 0.062 | 15.102 | 766 | 172 | 191 | GCA_001750085.1 | (48) |
| Glycine max (soybean) | Plantae | 11 107 | 995.27 | 47 064 | 38 274 | 0.236 | 11.160 | 6559 | 1295 | 3253 | GCF_000004515.6 | (49) |
| Gonium pectorale (green alga) | Plantae | 1028 | 148.81 | 16 290 | 2669 | 0.063 | 6.908 | 719 | 143 | 166 | GCA_001584585.1 | (50) |
| Musa acuminata (dwarf banana) | Plantae | 5489 | 461.54 | 22 177 | 20 934 | 0.179 | 11.893 | 2633 | 1083 | 1773 | GCF_000313855.2 | (51) |
| Oryza sativa (rice) | Plantae | 4531 | 386.49 | 28 735 | 14 704 | 0.158 | 11.723 | 2641 | 812 | 1078 | GCF_001433935.1 | (52) |
| Prunus persica (peach) | Plantae | 3454 | 220.9 | 23 133 | 11 210 | 0.149 | 15.636 | 2013 | 611 | 830 | GCF_000346465.2 | (53) |
| Solanum lycopersicum (tomato) | Plantae | 4144 | 809.18 | 25 612 | 13 711 | 0.162 | 5.121 | 2345 | 768 | 1031 | GCF_000188115.4 | (54) |
| Solanum tuberosum (potato) | Plantae | 4733 | 768.2 | 28 407 | 15 926 | 0.167 | 6.161 | 2647 | 880 | 1206 | GCF_000226075.1 | (55) |
| Theobroma cacao (cacao) | Plantae | 3074 | 335.44 | 21 517 | 9933 | 0.143 | 9.164 | 1775 | 554 | 745 | GCF_000208745.1 | (56) |
| Vitis vinifera (wine grape) | Plantae | 4039 | 427.21 | 25 830 | 13 613 | 0.156 | 9.454 | 2293 | 722 | 1024 | GCF_000003745.3 | (57) |
| Volvox carteri (green alga) | Plantae | 863 | 137.68 | 14 436 | 2600 | 0.061 | 6.268 | 509 | 152 | 202 | GCA_000143455.1 | (58) |
| Zea mays (Maize) | Plantae | 6801 | 2191.6 | 34 328 | 22 499 | 0.198 | 3.103 | 3910 | 1146 | 1745 | GCF_902167145.1 | (59) |
2-group HSDs refers to the number of curated HSD groups with only two gene copies.
Accession numbers are from the US NCBI GenBank assembly accession.
Summary statistics of the curated HSD groups in the selected genomes from HSDatabase
| Species name (common name) . | Classification . | Curated HSD groups # . | Genome size (Mb) . | No. of considered genes . | Gene copies . | HSDs/genes . | HSDs/Mb . | 2-group HSDsa . | 3-group HSDs . | ≥4-group HSDs . | Genome assembly accession numberb . | Ref . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ailuropoda melanoleuca (giant panda) | Animalia | 2534 | 2371.8 | 22 450 | 8091 | 0.113 | 1.068 | 1570 | 440 | 524 | GCF_002007445.1 | (23) |
| Bos taurus (cattle) | Animalia | 2238 | 2667.85 | 21 036 | 7399 | 0.106 | 0.839 | 1433 | 433 | 372 | GCF_002263795.1 | (24) |
| Canis lupus familiaris (dog) | Animalia | 2372 | 2344.09 | 20 945 | 7314 | 0.113 | 1.012 | 1541 | 434 | 397 | GCF_014441545.1 | (25) |
| Danio rerio (zebrafish) | Animalia | 3799 | 1405.1 | 26 439 | 11 930 | 0.144 | 2.704 | 2263 | 689 | 847 | GCF_000002035.6 | (26) |
| Drosophila melanogaster (fruit fly) | Animalia | 782 | 138.93 | 13 739 | 2230 | 0.057 | 5.629 | 557 | 118 | 107 | GCF_000001215.4 | (27) |
| Equus caballus (horse) | Animalia | 2403 | 2474.93 | 21 126 | 7499 | 0.114 | 0.971 | 1509 | 468 | 426 | GCF_002863925.1 | (28) |
| Felis catus (domestic cat) | Animalia | 2167 | 2493.14 | 20 452 | 6691 | 0.106 | 0.869 | 1409 | 396 | 362 | GCF_018350175.1 | (29) |
| Gadus morhua (Atlantic cod) | Animalia | 3269 | 627.04 | 23 482 | 9757 | 0.139 | 5.213 | 1969 | 633 | 667 | GCF_902167405.1 | (30) |
| Gallus gallus (chicken) | Animalia | 2026 | 1037.17 | 17 797 | 6092 | 0.114 | 1.953 | 1323 | 374 | 329 | GCF_016699485.2 | (31) |
| Gorilla gorilla (western gorilla) | Animalia | 2335 | 3063.36 | 20 632 | 6683 | 0.113 | 0.762 | 1531 | 434 | 370 | GCF_008122165.1 | (32) |
| Homo sapiens (human) | Animalia | 2178 | 2864.14 | 19 531 | 6352 | 0.112 | 0.760 | 1442 | 395 | 341 | GCF_000001405.39 | (33) |
| Hypsibius dujardini (waterbear) | Animalia | 2290 | 182.15 | 20 853 | 5710 | 0.110 | 12.572 | 1618 | 366 | 306 | GCA_002082055.1 | (34) |
| Loxodonta africana (African savanna elephant) | Animalia | 2031 | 3196.74 | 21 094 | 7371 | 0.096 | 0.635 | 1327 | 360 | 344 | GCF_000001905.1 | (35) |
| Meleagris gallopavo (turkey) | Animalia | 1486 | 1058.64 | 17 974 | 3968 | 0.083 | 1.404 | 1061 | 240 | 185 | GCF_000146605.3 | (36) |
| Mus musculus (house mouse) | Animalia | 2402 | 2588.62 | 30 736 | 8855 | 0.108 | 0.928 | 1480 | 459 | 463 | GCF_000001635.27 | (37) |
| Rattus norvegicus (Norway rat) | Animalia | 2434 | 2647.92 | 22 219 | 8757 | 0.110 | 0.919 | 1478 | 471 | 485 | GCF_015227675.2 | (38) |
| Saccharomyces cerevisiae (yeast) | Fungi | 397 | 11.83 | 6002 | 1006 | 0.073 | 33.559 | 331 | 41 | 25 | GCA_003086655.1 | (39) |
| Arabidopsis lyrata | Plantae | 5302 | 202.97 | 29 817 | 16 901 | 0.178 | 26.122 | 3104 | 985 | 1213 | GCF_000004255.2 | (40) |
| Arabidopsis thaliana (thale cress) | Plantae | 4428 | 119.75 | 27 560 | 14 225 | 0.161 | 36.977 | 2630 | 793 | 1005 | GCF_000001735.4 | (41) |
| Brassica oleracea (wild cabbage) | Plantae | 8918 | 529.92 | 44 382 | 30 511 | 0.201 | 16.829 | 4565 | 1841 | 2512 | GCF_000695525.1 | (42) |
| Carica papaya (papaya) | Plantae | 2094 | 360.63 | 18 126 | 6311 | 0.116 | 5.807 | 1299 | 374 | 421 | GCF_000150535.2 | (43) |
| Chlamydomonas eustigma (green alga) | Plantae | 966 | 66.63 | 14 161 | 2199 | 0.068 | 14.498 | 778 | 109 | 79 | GCA_002335675.1 | (20) |
| Chlamydomonas reinhardtii (green alga) | Plantae | 1129 | 111.11 | 19 870 | 3160 | 0.064 | 10.161 | 740 | 187 | 202 | GCF_000002595.2 | (44) |
| Chlamydomonas sp. ICE-L | Plantae | 1540 | 541.86 | 17 731 | 3853 | 0.078 | 2.842 | 1139 | 224 | 177 | GCA_013435795.1 | (21) |
| Chlamydomonas sp. UWO 241 (green alga) | Plantae | 1112 | 211.64 | 16 018 | 3282 | 0.068 | 5.254 | 741 | 139 | 232 | GCA_016618255.1 | (6) |
| Coccomyxa subellipsoidea C-169 (green alga) | Plantae | 360 | 48.83 | 9839 | 1015 | 0.037 | 7.373 | 281 | 43 | 36 | GCA_000258705.1 | (45) |
| Cucumis sativus (cucumber) | Plantae | 2891 | 240.99 | 20 038 | 9558 | 0.144 | 11.996 | 1655 | 532 | 704 | GCF_000004075.3 | (46) |
| Dunaliella salina (green alga) | Plantae | 1589 | 343.7 | 18 740 | 3859 | 0.095 | 4.623 | 1227 | 194 | 168 | GCA_002284615.2 | (47) |
| Fragilariopsis cylindrus (diatom) | Stramenopila | 1129 | 74.76 | 18 111 | 3192 | 0.062 | 15.102 | 766 | 172 | 191 | GCA_001750085.1 | (48) |
| Glycine max (soybean) | Plantae | 11 107 | 995.27 | 47 064 | 38 274 | 0.236 | 11.160 | 6559 | 1295 | 3253 | GCF_000004515.6 | (49) |
| Gonium pectorale (green alga) | Plantae | 1028 | 148.81 | 16 290 | 2669 | 0.063 | 6.908 | 719 | 143 | 166 | GCA_001584585.1 | (50) |
| Musa acuminata (dwarf banana) | Plantae | 5489 | 461.54 | 22 177 | 20 934 | 0.179 | 11.893 | 2633 | 1083 | 1773 | GCF_000313855.2 | (51) |
| Oryza sativa (rice) | Plantae | 4531 | 386.49 | 28 735 | 14 704 | 0.158 | 11.723 | 2641 | 812 | 1078 | GCF_001433935.1 | (52) |
| Prunus persica (peach) | Plantae | 3454 | 220.9 | 23 133 | 11 210 | 0.149 | 15.636 | 2013 | 611 | 830 | GCF_000346465.2 | (53) |
| Solanum lycopersicum (tomato) | Plantae | 4144 | 809.18 | 25 612 | 13 711 | 0.162 | 5.121 | 2345 | 768 | 1031 | GCF_000188115.4 | (54) |
| Solanum tuberosum (potato) | Plantae | 4733 | 768.2 | 28 407 | 15 926 | 0.167 | 6.161 | 2647 | 880 | 1206 | GCF_000226075.1 | (55) |
| Theobroma cacao (cacao) | Plantae | 3074 | 335.44 | 21 517 | 9933 | 0.143 | 9.164 | 1775 | 554 | 745 | GCF_000208745.1 | (56) |
| Vitis vinifera (wine grape) | Plantae | 4039 | 427.21 | 25 830 | 13 613 | 0.156 | 9.454 | 2293 | 722 | 1024 | GCF_000003745.3 | (57) |
| Volvox carteri (green alga) | Plantae | 863 | 137.68 | 14 436 | 2600 | 0.061 | 6.268 | 509 | 152 | 202 | GCA_000143455.1 | (58) |
| Zea mays (Maize) | Plantae | 6801 | 2191.6 | 34 328 | 22 499 | 0.198 | 3.103 | 3910 | 1146 | 1745 | GCF_902167145.1 | (59) |
| Species name (common name) . | Classification . | Curated HSD groups # . | Genome size (Mb) . | No. of considered genes . | Gene copies . | HSDs/genes . | HSDs/Mb . | 2-group HSDsa . | 3-group HSDs . | ≥4-group HSDs . | Genome assembly accession numberb . | Ref . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ailuropoda melanoleuca (giant panda) | Animalia | 2534 | 2371.8 | 22 450 | 8091 | 0.113 | 1.068 | 1570 | 440 | 524 | GCF_002007445.1 | (23) |
| Bos taurus (cattle) | Animalia | 2238 | 2667.85 | 21 036 | 7399 | 0.106 | 0.839 | 1433 | 433 | 372 | GCF_002263795.1 | (24) |
| Canis lupus familiaris (dog) | Animalia | 2372 | 2344.09 | 20 945 | 7314 | 0.113 | 1.012 | 1541 | 434 | 397 | GCF_014441545.1 | (25) |
| Danio rerio (zebrafish) | Animalia | 3799 | 1405.1 | 26 439 | 11 930 | 0.144 | 2.704 | 2263 | 689 | 847 | GCF_000002035.6 | (26) |
| Drosophila melanogaster (fruit fly) | Animalia | 782 | 138.93 | 13 739 | 2230 | 0.057 | 5.629 | 557 | 118 | 107 | GCF_000001215.4 | (27) |
| Equus caballus (horse) | Animalia | 2403 | 2474.93 | 21 126 | 7499 | 0.114 | 0.971 | 1509 | 468 | 426 | GCF_002863925.1 | (28) |
| Felis catus (domestic cat) | Animalia | 2167 | 2493.14 | 20 452 | 6691 | 0.106 | 0.869 | 1409 | 396 | 362 | GCF_018350175.1 | (29) |
| Gadus morhua (Atlantic cod) | Animalia | 3269 | 627.04 | 23 482 | 9757 | 0.139 | 5.213 | 1969 | 633 | 667 | GCF_902167405.1 | (30) |
| Gallus gallus (chicken) | Animalia | 2026 | 1037.17 | 17 797 | 6092 | 0.114 | 1.953 | 1323 | 374 | 329 | GCF_016699485.2 | (31) |
| Gorilla gorilla (western gorilla) | Animalia | 2335 | 3063.36 | 20 632 | 6683 | 0.113 | 0.762 | 1531 | 434 | 370 | GCF_008122165.1 | (32) |
| Homo sapiens (human) | Animalia | 2178 | 2864.14 | 19 531 | 6352 | 0.112 | 0.760 | 1442 | 395 | 341 | GCF_000001405.39 | (33) |
| Hypsibius dujardini (waterbear) | Animalia | 2290 | 182.15 | 20 853 | 5710 | 0.110 | 12.572 | 1618 | 366 | 306 | GCA_002082055.1 | (34) |
| Loxodonta africana (African savanna elephant) | Animalia | 2031 | 3196.74 | 21 094 | 7371 | 0.096 | 0.635 | 1327 | 360 | 344 | GCF_000001905.1 | (35) |
| Meleagris gallopavo (turkey) | Animalia | 1486 | 1058.64 | 17 974 | 3968 | 0.083 | 1.404 | 1061 | 240 | 185 | GCF_000146605.3 | (36) |
| Mus musculus (house mouse) | Animalia | 2402 | 2588.62 | 30 736 | 8855 | 0.108 | 0.928 | 1480 | 459 | 463 | GCF_000001635.27 | (37) |
| Rattus norvegicus (Norway rat) | Animalia | 2434 | 2647.92 | 22 219 | 8757 | 0.110 | 0.919 | 1478 | 471 | 485 | GCF_015227675.2 | (38) |
| Saccharomyces cerevisiae (yeast) | Fungi | 397 | 11.83 | 6002 | 1006 | 0.073 | 33.559 | 331 | 41 | 25 | GCA_003086655.1 | (39) |
| Arabidopsis lyrata | Plantae | 5302 | 202.97 | 29 817 | 16 901 | 0.178 | 26.122 | 3104 | 985 | 1213 | GCF_000004255.2 | (40) |
| Arabidopsis thaliana (thale cress) | Plantae | 4428 | 119.75 | 27 560 | 14 225 | 0.161 | 36.977 | 2630 | 793 | 1005 | GCF_000001735.4 | (41) |
| Brassica oleracea (wild cabbage) | Plantae | 8918 | 529.92 | 44 382 | 30 511 | 0.201 | 16.829 | 4565 | 1841 | 2512 | GCF_000695525.1 | (42) |
| Carica papaya (papaya) | Plantae | 2094 | 360.63 | 18 126 | 6311 | 0.116 | 5.807 | 1299 | 374 | 421 | GCF_000150535.2 | (43) |
| Chlamydomonas eustigma (green alga) | Plantae | 966 | 66.63 | 14 161 | 2199 | 0.068 | 14.498 | 778 | 109 | 79 | GCA_002335675.1 | (20) |
| Chlamydomonas reinhardtii (green alga) | Plantae | 1129 | 111.11 | 19 870 | 3160 | 0.064 | 10.161 | 740 | 187 | 202 | GCF_000002595.2 | (44) |
| Chlamydomonas sp. ICE-L | Plantae | 1540 | 541.86 | 17 731 | 3853 | 0.078 | 2.842 | 1139 | 224 | 177 | GCA_013435795.1 | (21) |
| Chlamydomonas sp. UWO 241 (green alga) | Plantae | 1112 | 211.64 | 16 018 | 3282 | 0.068 | 5.254 | 741 | 139 | 232 | GCA_016618255.1 | (6) |
| Coccomyxa subellipsoidea C-169 (green alga) | Plantae | 360 | 48.83 | 9839 | 1015 | 0.037 | 7.373 | 281 | 43 | 36 | GCA_000258705.1 | (45) |
| Cucumis sativus (cucumber) | Plantae | 2891 | 240.99 | 20 038 | 9558 | 0.144 | 11.996 | 1655 | 532 | 704 | GCF_000004075.3 | (46) |
| Dunaliella salina (green alga) | Plantae | 1589 | 343.7 | 18 740 | 3859 | 0.095 | 4.623 | 1227 | 194 | 168 | GCA_002284615.2 | (47) |
| Fragilariopsis cylindrus (diatom) | Stramenopila | 1129 | 74.76 | 18 111 | 3192 | 0.062 | 15.102 | 766 | 172 | 191 | GCA_001750085.1 | (48) |
| Glycine max (soybean) | Plantae | 11 107 | 995.27 | 47 064 | 38 274 | 0.236 | 11.160 | 6559 | 1295 | 3253 | GCF_000004515.6 | (49) |
| Gonium pectorale (green alga) | Plantae | 1028 | 148.81 | 16 290 | 2669 | 0.063 | 6.908 | 719 | 143 | 166 | GCA_001584585.1 | (50) |
| Musa acuminata (dwarf banana) | Plantae | 5489 | 461.54 | 22 177 | 20 934 | 0.179 | 11.893 | 2633 | 1083 | 1773 | GCF_000313855.2 | (51) |
| Oryza sativa (rice) | Plantae | 4531 | 386.49 | 28 735 | 14 704 | 0.158 | 11.723 | 2641 | 812 | 1078 | GCF_001433935.1 | (52) |
| Prunus persica (peach) | Plantae | 3454 | 220.9 | 23 133 | 11 210 | 0.149 | 15.636 | 2013 | 611 | 830 | GCF_000346465.2 | (53) |
| Solanum lycopersicum (tomato) | Plantae | 4144 | 809.18 | 25 612 | 13 711 | 0.162 | 5.121 | 2345 | 768 | 1031 | GCF_000188115.4 | (54) |
| Solanum tuberosum (potato) | Plantae | 4733 | 768.2 | 28 407 | 15 926 | 0.167 | 6.161 | 2647 | 880 | 1206 | GCF_000226075.1 | (55) |
| Theobroma cacao (cacao) | Plantae | 3074 | 335.44 | 21 517 | 9933 | 0.143 | 9.164 | 1775 | 554 | 745 | GCF_000208745.1 | (56) |
| Vitis vinifera (wine grape) | Plantae | 4039 | 427.21 | 25 830 | 13 613 | 0.156 | 9.454 | 2293 | 722 | 1024 | GCF_000003745.3 | (57) |
| Volvox carteri (green alga) | Plantae | 863 | 137.68 | 14 436 | 2600 | 0.061 | 6.268 | 509 | 152 | 202 | GCA_000143455.1 | (58) |
| Zea mays (Maize) | Plantae | 6801 | 2191.6 | 34 328 | 22 499 | 0.198 | 3.103 | 3910 | 1146 | 1745 | GCF_902167145.1 | (59) |
2-group HSDs refers to the number of curated HSD groups with only two gene copies.
Accession numbers are from the US NCBI GenBank assembly accession.
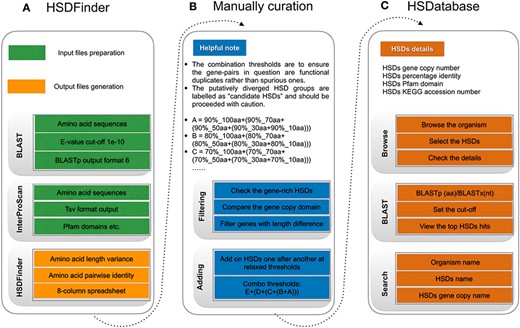
The workflow of HSDatabase. (A) Steps for using HSDFinder to collect candidate HSDs. (B) Manual curation of HSDs via filtering and adding new HSD candidates prior to being deposited into HSDatabase. (C) Steps of accessing HSD data in HSDatabase, including browsing via organism name, blasting query sequences against the database and searching through the HSD and gene copy IDs.
Database curation
Prior to uploading data into HSDatabase, we curated HSD candidates by filtering for redundancy and adding the newly curated HSDs (Figure 2B). For genes that have alternative protein products, we selected the longest gene isoform to reduce redundancy. Since highly similar gene copies are grouped together as HSDs based on a simple transitive link between the remaining genes (14), it is possible for some highly duplicated genes to form mega HSD groups with varied gene copy lengths, especially those encoding histones, ribosomal proteins or retro-transcriptases. Moreover, some gene copies might appear multiple times causing redundancy among different HSD groups, which is because the BLAST algorithm limits the maximum target hits by default. In these cases, we manually curated the HSD groups, minimizing redundant gene copies.
Database implementation
The database was built with the Django 3.0.5 web framework (https://www.djangoproject.com/), and all data were stored in an SQLite 3.36.0 database (https://www.sqlite.org/index.html) on an Amazon web server. Webpage templates used Bootstrap framework (https://getbootstrap.com/), D3.js (https://d3js.org), jQuery (http://jquery.com) and Bootstrap Table (https://bootstrap-table.com/) libraries to establish a user-friendly, front-end interface. On the browse page, NCBI’s Sequence Viewer 3.44.0 (https://www.ncbi.nlm.nih.gov/projects/sviewer/) was employed to build a fast and scalable genome browser.
Results and discussion
Database content and analysis
HSDatabase was built using a relational database (MySQL) allowing the rapid retrieval of data and making resources easily maintainable. One entry corresponds to one eukaryote genome. The genomes can be accessed via the organism table or the taxonomic tree. The genome entry is then split into various subcategories of HSD entries. Database access is via a web interface based on python script and provides various ways to search for HSD entries, including species name, unique HSD IDs and gene copy IDs.
Using HSDFinder (15), we collected and curated 117 864 HSDs (representing 379 844 gene copies) from 40 well-assembled nuclear genomes of diverse model species (Table 1). Various green algae were included because of our specific interest in algal genomics and also because of their relatively modest genome sizes and penchant for gene duplications. For example, the acidophilic green alga Chlamydomonas eustigma is known to have large numbers of gene duplicates in its nuclear genome, including 10 gene copies for arsenate reductase and 20 for glutaredoxin (20). Similarly, the psychrophilic green alga Chlamydomonas sp. ICE-L contains multiple copies of genes encoding carotene biosynthesis-related protein and Lhc-like protein (Lhl4) (21). These data are consistent with our identification of large numbers of HSDs in C. eustigma (276) and ICE-L (265) (Table 1), suggesting a potential adaptative role of gene duplication under different extreme environmental conditions.
Compared to algae, the investigated land plants had higher detected numbers of HSDs as well as larger ratios of HSDs/Mb and HSDs/genes (Table 1). For example, the HSDs/Mb values for Arabidopsis lyrata and A. thaliana are 26 and 37, respectively, whereas the average HSDs/Mb value among selected green algae is 8.2. Compared to algae and land plants, the HSDs/Mb values in animals are generally quite low with the exception of Hypsibius dujardini (13.6) and D. melanogaster (5.6). Two-group HSDs (i.e. HSDs containing two gene copies) represent the majority (>50%) of total HSDs for all explored species.
As for the associated functions of the detected HSDs, three green algal species with relatively large values of HSDs/genes were compared previously. These algae can survive various extreme environmental conditions and include the Antarctic psychrophilic green algae UWO241 (0.068) and ICE-L (0.078) and the acidophilic C. eustigma (0.068) (Table 1). The identified duplicates are involved in a diversity of cellular pathways, including gene expression, cell growth, membrane transport and energy metabolism, but also include ribosomal proteins (6, 14). Although HSDs for protein translation, DNA packaging and photosynthesis are particularly prevalent, around 30% of the HSDs are hypothetical proteins without any Pfam domains.
Database composition and usage
Information about specific HSDs and their associated gene copies for a given species can be obtained through the ‘Browse’ and ‘Search’ tabs, which are located on the menu bar at the top of the page, or using nucleotide/amino acid sequences as queries to search against the database via BLAST (i.e. BLASTP or BLASTX). To categorize duplicated genes into their functional categories, KEGG pathway schematics are available for each species.
Browse
By selecting the ‘Browse’ option from the main menu, users are offered three ways to explore their species of interest. First, they can simply click the organism name on the taxonomic tree containing the 40 species. Secondly, users can select the ‘Plantae and Stramenopila’ or ‘Animalia and Fungi’ tabs (Figure 3A), which contain 23 and 17 species, respectively. Selecting a tab takes users to a summary table that contains the organism names, number of HSDs, species background information, GenBank accession links to genome assemblies, and reference links to PubMed.
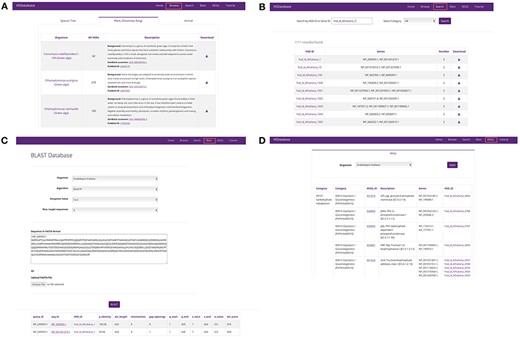
Screenshots of the HSDatabase interface. There are four main functions in the menu page: (A) Browse the database via species entries; (B) search the database via the HSDatabase unique ID (e.g. hsd_id_Athaliana_1) or gene ID (e.g. NP_200993.1); (C) use BLAST to search the database via amino acid sequence in FASTA format; and (D) categorize the gene copies and HSDs under the KEGG pathway functional categories.
Selecting a specific species from the browse page leads to the respective HSDs summary page (Figure 4A), which gives data on the total number of HSDs, unique HSD IDs, gene copy GenBank IDs and number of gene copies; it also provides access to the data download function. Choosing one of the HSD ID entries, for example, brings up a page containing information and features of a detected HSD, including the associated gene copies for a unique HSD as well as the GenBank link, the sequence length, the Pfam domain ID/description and the InterPro database ID/description (Figure 4B). Clicking on the ‘genome browser’ tab allows for the visualization of a specific gene copy through the built-in NCBI genome browser (Figure 4C). The ‘FASTA sequence’ tab provides the option to download the sequence data (Figure 4D) and the ‘alignments and identity%’ tab brings up the gene copy alignment and percentage identity matrix created by the built-in Clustal v2.1 tool (22) (Figure 4E).
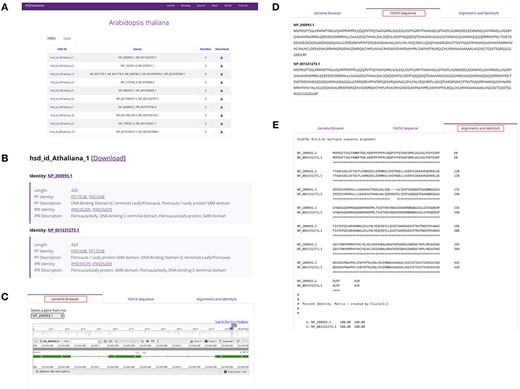
Summary of database information for a selected species. (A) HSDs collected in a table for a specific species. (B) Basic information of the unique HSD ID, gene copy ID and the associated links to Pfam domains and InterPro databases. (C) Linking gene copies to the genome browser. (D) The FASTA sequence downloads of gene copies. (E) Alignments and percentage sequence identities of gene copies.
Search
Through the search option from the main menu users can search unique HSD IDs or gene copy IDs against the database (Figure 3B); they can also set the selection categories to limit the search results, which can improve search efficiency. After activation of the search button, 30 results per page are displayed (Figure 3B) in a four-column table, including HSD name, gene copy name, number of gene copies and the external download link to the output data (tab-delimited file). Users can navigate through the results page or download specific HSD entries. As described in the Browse section, the data file includes various summary statistics on the HSDs (Figure 4B).
BLAST
The BLAST tool bar allows users to input a nucleotide or amino acid sequence (in FASTA format) and carry out a sequence similarity search using BLASTX or BLASTP. Users can specify the species against which the BLAST search will be performed. The E-value and maximum target sequence of results can also be adjusted, but all other parameters remain as default and cannot be changed (Figure 3C). The BLAST search output result is in the standard 13-column tabular format, including the linkable query sequence ID and HSD ID, percentage identity, aligned length and all other BLAST tabular output values. The most similar sequences are arranged at the top.
KEGG
The KEGG page contains details on the associated KEGG pathways of the HSD gene copies for the 40 species. To browse the data for a particular species, users can simply select the organism’s name. The 6-column table lists the gene copies and HSDs under KEGG functional categories (Figure 3D). Gene copies involved in the same KEGG pathway are detailed with the first KEGG category (e.g. Carbohydrate metabolism), then the secondary category (e.g. Glycolysis/Gluconeogenesis) and finally the KEGG pathway function description (e.g. ENO, eno; enolase). The KEGG ID (e.g. K01689) is linked to the external KEGG database, providing more detailed information.
Future direction and limitation
Now that HSDatabase is publicly available, the next step is to analyze duplicate genes across a broader range of species, which we plan on doing in the near future. Currently, the database includes a range of statistics (e.g. number of HSD per Mb), but we hope to add additional data in the coming years, including information on differential expression levels among duplicates, for instance, as well as data on rates of synonymous and nonsynonymous substitutions (dN/dS rates). The biggest challenge moving forward will be determining an appropriate threshold for accurately predicting HSDs. As research on gene duplicates improves, we may need to adjust the metrics (e.g. amino acid pairwise identity and amino acid aligned length variance) to find as many bona fide HSDs as possible.
Presently, there is no standard golden cut-off for identifying HSDs and there might never be one as there a multitude of forces, including lineage/genomic specific ones, that can impact the accuracy of the identification metrics. This is why users can employ different parameters in the HSDFinder tool (from 30 to 100% amino acid pairwise identity and from within 0 to 100 amino acid aligned length variance). In our case, we used a series of combination thresholds to curate the HSDs in HSDatabase. But due to the limitations of this strategy, there are some large groups of HSD candidates in the database that likely diverged in function from one another and, thus, are not inducing a gene dosage benefit. In the database, we have labeled these putatively diverged HSD groups as ‘candidate HSDs’ and have added a warning note that users should proceed with caution when working with these datasets. In the future, our goal is to guide users to species-specific thresholds and deposit more diverse eukaryotic species into the database.
Conclusions
With the decreasing cost of next-generation sequencing, biologists are dealing with ever larger amounts of data. However, many bioinformatics software suites require considerable knowledge of computer scripting and microprogramming. To facilitate the understanding and analysis of gene duplication in nuclear genomes, we developed HSDatabase, which currently contains 117 864 HSDs from 40 well-assembled eukaryotic genomes. In conjunction with HSDatabase, we designed HSDFinder, which can efficiently identify duplicated genes from unannotated genome sequences by integrating the results from InterProScan and KEGG. HSDatabase aims to become a useful platform for the identification and comprehensive analysis of HSDs in eukaryotic genomes, which could aid research into the mechanisms driving genome adaptation. In the future, the database will be updated by incorporating advancements in the field of gene duplication.
Supplementary data
Supplementary data are available at Database Online.
Acknowledgements
We want to especially thank the editors and reviewers for their professional comments that greatly improved this manuscript.
Funding
Discovery Grants from the Natural Sciences and Engineering Research Council of Canada.
Conflict of interest
None declared.
Data availability
The datasets of eukaryotes supporting the conclusions of this article are available from Joint Genome Institute (JGI) (https://phytozome.jgi.doe.gov/pz/portal.html) or National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) database.
Author contributions
The study was conceptualized by X.Z. and D.R.S. The data were analyzed by X.Z., and Y.N.H. implemented the HSDatabase website. X.Z. and D.R.S. drafted the manuscript, and all authors commented to produce the manuscript for peer review.



