-
PDF
- Split View
-
Views
-
Cite
Cite
Gjorgjina Cenikj, Eva Valenčič, Gordana Ispirova, Matevž Ogrinc, Riste Stojanov, Peter Korošec, Ermanno Cavalli, Barbara Koroušić Seljak, Tome Eftimov, CafeteriaSA corpus: scientific abstracts annotated across different food semantic resources, Database, Volume 2022, 2022, baac107, https://doi.org/10.1093/database/baac107
Close - Share Icon Share
Abstract
In the last decades, a great amount of work has been done in predictive modeling of issues related to human and environmental health. Resolution of issues related to healthcare is made possible by the existence of several biomedical vocabularies and standards, which play a crucial role in understanding the health information, together with a large amount of health-related data. However, despite a large number of available resources and work done in the health and environmental domains, there is a lack of semantic resources that can be utilized in the food and nutrition domain, as well as their interconnections. For this purpose, in a European Food Safety Authority–funded project CAFETERIA, we have developed the first annotated corpus of 500 scientific abstracts that consists of 6407 annotated food entities with regard to Hansard taxonomy, 4299 for FoodOn and 3623 for SNOMED-CT. The CafeteriaSA corpus will enable the further development of natural language processing methods for food information extraction from textual data that will allow extracting food information from scientific textual data.
Database URL: https://zenodo.org/record/6683798#.Y49wIezMJJF
Introduction
Nowadays, there are many scientific publications that contain valuable information about food and nutrition. This information needs to be systematically reviewed in order to find answers to open research questions, which requires an investigation of interactions between food, as one of the main environmental factors, and other health-related factors, such as diseases, treatments and drugs. However, it is difficult and time-consuming to keep up with the new insights (knowledge) that are being published every day with new scientific publications. For this purpose, natural language processing (NLP) methods can facilitate the speedup and automation of the process of extracting relevant information (1). In order to be able to train such methods to learn models that are able to do this, we need a gold standard of annotated corpora, which consists of scientific abstracts that are already annotated with concepts and entities (related, e.g. to diseases, drugs and treatments in the biomedical domain) of interest.
The NLP task for identifying the entities mentioned in unstructured textual data and further classifying them into predefined categories (e.g. diseases, drugs, treatments and genes) is known as named-entity recognition (NER) (2). Several types of NER methods exist based on the methodology they are using:
Dictionary-based methods—they extract the entities that exist in a predefined dictionary that is used as a lookup table for searching (3). Their performance depends on the comprehensive coverage of the dictionary.
Rule-based methods—they are based on dictionaries used for searching combined with handwritten rules that describe the characteristics of the entities of interest (4, 5). Their weakness is the time required to write domain-specific rules; however, they are still beneficial when an annotated corpus is not available, especially in a low-resource domain.
Corpus-based methods—they require an annotated corpus that is used with some supervised machine learning (ML) method to learn a classification model (6, 7). The classes are represented as tags in the inside–outside–beginning (IOB) tagging scheme, where the goal is to determine whether each token (i.e. word) in the text is outside, inside or at the beginning of an entity of interest. Their strength is that they provide really robust results; however, it comes with the cost that large annotated corpora should be available.
Methods based on active learning—they train a semi-supervised model that starts with a small amount of annotated data and further interacts with a user to query for new annotations that are used to iteratively improve the model’s performance (8).
A lot of publications already exist in the biomedical domain (9–13), where the focus is on the development of annotated corpora that will further allow learning of biomedical NER methods. This is also supported by biomedical shared workshops such as BioCreative (14, 15), i2b2 (16) and BioNLP (17), where annotated corpora from the biomedical domain are published every year with different scopes that help the community develop robust NER methods. A nice overview of the existing corpora in the biomedical domain together with open challenges is presented in reference (18), from where it can be seen that in the last 15 years the focus is on extracting genes, proteins, chemicals, diseases, drugs and treatments. It is clear that food is not among them, and thus, there is a research gap and limited resources that can be utilized for the extraction of food entities that are needed to trace and address different applications of food safety.
Unlike the large number of semantic resources that are available in the biomedical domain, the food domain is still low resourced. There exist several rule-based NER methods such as drNER (4) and FoodIE (5) developed to help the extraction of food entities. drNER can extract food entities based on external dictionaries combined with rules based on Boolean algebra, while FoodIE uses an external semantic tagger and combines the semantic tags together with rules from computational linguistics. FoodIE allowed the creation of the first annotated corpus defining food entities, known as FoodBase (19), which consists of 1000 recipes represented with their textual descriptions (in English), where the food entities are annotated with food semantic tags from the Hansard taxonomy.
Recently, two corpus-based FoodNER methods have been proposed, known as Bidirectional Long Short-Term Memory for Food Named Entity Recognition (BuTTeR) (20) and FoodNER (21), both trained using the FoodBase corpus as training data. BuTTeR is trained using bidirectional long–short-term memory network in combination with a conditional random field in order to distinguish between food and non-food entities. However, FoodNER involves several different models, where Bidirectional Encoder Representations from Transformers (BERT) (22), Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT) (23) and A Robustly Optimized BERT Pretraining Approach (24) have been trained to extract the food entities and also assigned them food semantic tags from several semantic resources involving Hansard (25, 26), FoodOn (27) and SNOMED-CT (28). Even though both methods achieve high performance when applied to text from the same domain as the one they were trained on, i.e. recipe instructions, they cannot generalize to the task of food entity extraction from scientific abstracts. This is primarily due to scientific text having a vastly different writing style and contents compared to recipe text, as well as the fact that scientific articles related to food typically contain other entities that are not present in recipe data, such as chemicals, drugs and diseases. Some of the errors produced by FoodNER models trained on recipe text and applied to scientific text include false positives, where the model extracts the aforementioned types of entities as foods since they were not present in the training data, i.e. recipe text.
Recently, classical ML models were also used to train FoodNER methods utilizing the FoodBase corpus (29). The models reported in this study can also recognize nutrient and chemical entities that are not possible since the FoodBase corpus does not consist of such kinds of entities. Furthermore, they augmented the FoodBase with already existing scientific abstracts that consist of nutrient and chemical entities. The results from scientific abstracts are only about chemical and nutrition entities and not food entities.
To support the generalization of FoodNER methods, the European Food Safety Authority (EFSA) funds the project CAFETERIA for developing semantic resources that will further allow the development of FoodNER methods. As a part of it, we have developed the first annotated corpus of scientific text abstracts that contains annotations of food entities. We have called this corpus CafeteriaSA, which is a gold corpus of 500 annotated scientific abstracts available in the following four different versions depending on the semantic resource that is used for annotation: food vs. non-food, Hansard taxonomy, FoodOn and SNOMED-CT.
A part of the CAFETERIA project also involved the extension of the FoodBase corpus with semantic tags of the annotated mentions of food entities. As a result of this work, the CafeteriaFCD corpus (30) was produced, which is a version of the FoodBase corpus where the annotated food entities are linked to the Hansard taxonomy and the FoodOn and SNOMED-CT ontologies. The same methodology for automatic linking of the mentions of food entities in the text to the identifiers in the FoodOn and SNOMED-CT ontologies is used for adding the semantic tags to both the CafeteriaFCD corpus and the CafeteriaSA corpus (presented as a part of this work). However, the focus of these works differs in the fact that the CafeteriaFCD corpus is merely an extension of the FoodBase corpus, while here we present the entire process of annotating the CafeteriaSA corpus, which involves both the annotation of mentions of food entities in text and their automatic linking to the aforementioned resources.
The remainder of this paper is structured as follows: Materials and methods section explains the semantic resources utilized and the invented pipeline for the development of the CafeteriaSA corpus; in Results and discussion section, we present the coverage statistics of the developed corpus; finally, in Conclusions section, we conclude the paper, pointing out the possible applications for which such resources are required.
Materials and methods
In this section, we briefly describe the existing semantic resources utilized for the development of the CafeteriaSA corpus. For this purpose, semantic resources, such as Hansard taxonomy, FoodOn and SNOMED-CT, are explained, followed by two corpus-based FoodNER methods trained on the FoodBase corpus. Next, a recently proposed and published human–computer interaction (HCI) tool, known as FoodViz, is introduced. The FoodViz web-based tool is aimed to be used in the process of validating the semantic tags by domain experts. Finally, we provide a description of a pipeline developed for the creation of the CafeteriaSA corpus.
Food semantic resources
Hansard taxonomy
The Hansard taxonomy (25, 26) is a hierarchical organization of >8000 different semantic categories, where food and drink is one of the top-level categories, with food, production of food, farming and acquisition of animals for food and hunting as its immediate subcategories. Some food and drink categories are specified in many details (e.g. ‘vegetables’ or ‘drink’), while others are relatively broadly defined. This taxonomy is a useful linguistic resource compiled from transcribed speeches; however, its main gap is its sustainability.
FoodOn
FoodOn (27) is currently one of the strongest harmonized food ontologies, connecting a number of more specialized ontologies with the goal of eliminating the incompatibility and ambiguity of food references. It covers animal and plant food sources, terms related to cooking, packaging and preservation processes, as well as product-type schemes for the categorization of food products. The main feature of FoodOn is that it is developed by a global community of researchers and is in line with the widely accepted and used description and classification systems such as LanguaL (31) and FoodEx2 (32).
SNOMED-CT
SNOMED-CT (28) is a standardized, multilingual healthcare terminology that provides a consistent way to index and store clinical data. One of its primary uses is the representation of patient data in the form of electronic health records. The terminology includes relations between different body structures; organisms; substances; pharmaceutical products; physical objects; physical forces; specimens; symptoms; drugs; food; and surgical, therapeutic and diagnostic procedures.
Food named–entity recognition method
BuTTER
BuTTER (20) is the first corpus-based NER model in the food domain, trained on the FoodBase corpus. In order to identify food entities from raw text, it uses a neural network based on Bidirectional Long Short-Term Memory and Conditional Random Fields and pre-trained word embeddings. The BuTTER model achieves a macro-averaged F1 score of 0.94 for the extraction of food entities from recipes; however, it fails to generalize to scientific text.
FoodNER
FoodNER (21) is another corpus-based NER model, which performs fine-tuning of the transformer-based text representation models BERT (22) and BioBERT (23) on the FoodBase corpus. Apart from the NER task, the FoodNER model can also categorize the extracted entities and link them to the concepts in the FoodOn and SNOMED-CT ontologies and the Hansard taxonomy. The FoodNER model can identify food entities from recipes with a macro-averaged F1 score of 0.94. In the food entity linking tasks, it achieves macro-averaged scores in the range 0.73–0.78.
Human–computer annotation tool—FoodViz
In order for food experts to understand the links between different food semantic tags from different semantic resources and to make them familiar with the interoperability process using different standards, we have developed the FoodViz tool (33). The tool is an HCI tool implemented to present food annotation results from the existing ML models in conjunction with different food semantic data resources.
The FoodViz (http://foodviz.env4health.finki.ukim.mk/#/recipes) tool is a web-based application developed with React (https://reactjs.org/), served by a back-end application programming interface (API) developed in Flask (https://flask.palletsprojects.com/en/1.1.x/). It visualizes the recipes and annotations published in the FoodBase corpus. Its first appearance allows users to filter the recipes available in the FoodBase corpus by name or by the recipe category and to search in two different datasets: the curated recipes (i.e. where the food semantic tags for the recipes are manually corrected by domain experts after the automatic annotation with ML models) and uncurated (i.e. for recipes annotated using ML models, no domain expert validation is applied) recipes. The tool helps domain experts to understand the semantic resources together with NER methods. In addition, it allows them to remove the errors in the annotations provided by the ML models and also to add any missed entity that was not automatically recognized.
CAFETERIA annotation pipeline
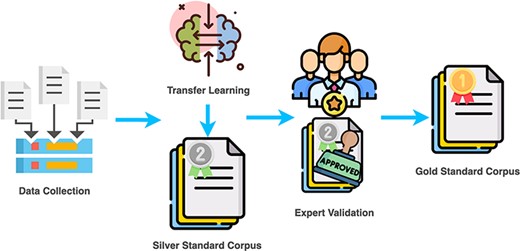
Next, an annotation pipeline, used to create the CafeteriaSA, will be explained in more detail. The pipeline is a synergy of the semantic resources mentioned earlier. It consists of four steps (see Figure 1): (i) collecting scientific abstract data, (ii) automatic annotation using the already developed food corpus–based NER methods with regard to the Hansard taxonomy—this step performs the generalization of the learned knowledge from another dataset that annotated recipe data, (iii) domain expert validation of the automatic annotations using the FoodViz tool and lastly (iv) alignment with other food semantic resources using the National Center for Biomedical Ontology (NCBO) annotator (34).
Step 1: Data collection
The abstracts of the scientific papers were collected from PubMed using Entrez Programming Utilities (35). The ESearch utility requires the definition of a search term and produces a set of unique identifiers of papers related to the search term. The EFetch API call can then be used to retrieve the paper data for each of the identifiers returned by the ESearch utility. Apart from the text of the paper abstract, the EFetch utility provides other information such as the title of the paper, the year the paper was published, the journal it was published in and MeSH terms of concepts that are discussed in the paper. We only included the scientific papers written in English. We do not pose a limitation on the time when the papers were published; however, the data collection process was executed in March 2021, so the scientific papers were published before this time.
In agreement with a domain expert from the team, the following 17 phrases were used as search terms for obtaining the initial set of abstracts related to food: asthma food, arthritis food, Parkinson disease food, bronchitis food, stroke food, food allergy, heart disease food, diabetes food, kidney stone food, anemia food, osteoporosis food, pneumonia food, Alzheimer food, skin disease food, tuberculosis food, hypertension food and influenza food. This resulted in 14 712 paper identifiers retrieved from the ESearch utility. For each identifier, the EFetch utility was used to retrieve the additional information for each paper.
For annotation, 500 scientific abstracts were selected. The selection was based on the number of food entities found in abstracts using a voting scheme (see Figure 2) and journals in which they were published. First, to ensure that abstracts contained sufficient food-relevant information, we have limited ourselves to abstracts that were identified to have at least eight food entities, in order to fully utilize the experts’ effort and not have them check abstracts in which no food entities were present. There were 1782 abstracts that fulfilled such criteria and were published in 686 journals. Second, journals of these abstracts were organized in a table, where for each journal we calculated the number of abstracts. Based on the consultation with domain experts, we have selected 500 abstracts from 47 journals covering different food safety subdomains. We have done this in order to increase the variability in the writing style and enable the annotation of diverse entities.
Step 2: Automatic annotation using corpus-based NER methods
To obtain food annotations of the selected abstracts, we used a voting scheme that combined the annotations of food entities produced by the corpus-based NER methods mentioned earlier, BuTTeR and FoodNER, together with two dictionary-based models using dictionaries containing food entities extracted from the FooDB database. We have decided to perform an ensemble to annotate the data since using all these resources separately leads to lower performance in the annotation. This is partly due to the fact that dictionary-based NER models (such as the ones based on FooDB) are completely dependent on the quality of the dictionary they are based on and thus require extensive dictionaries that provide a good coverage of the variations and synonyms of entity names to produce satisfying results. They are also prone to producing partial entity matches when an entity consisting of several words is not present in the dictionary, but one of its constituent words is. On the other hand, corpus-based NER models are highly dependent on the corpus they were trained on, and since BuTTER and FoodNER are trained on recipe text, which is vastly different from scientific text, they fail to generalize to scientific text.
The lack of a scientific corpus annotated with food entities did not allow us to evaluate and compare the existing FoodNER methods on scientific text, so we opted for a synergy of several NER methods integrated into a voting scheme since such an approach has shown to produce better results in some of our previous experiments. In addition, the initial automatic labeling of the scientific abstracts was only meant to speed up the process of manual curation. Our ultimate goal was not to find the best NER model but to create a novel corpus annotated with food entities, which will enable a fair evaluation of the existing methods and the generation of novel ones.
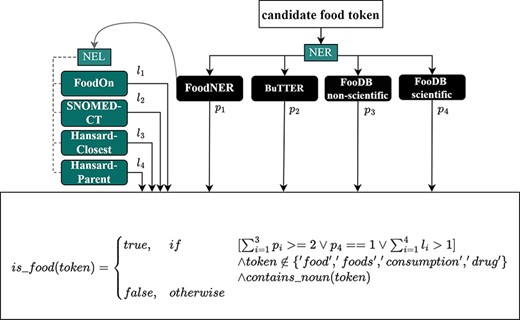
Figure 2 presents the voting scheme used for FoodNER. The voting scheme combines the annotations of the BuTTER and FoodNER models with annotations of two dictionary-based methods, which we refer to FooDB scientific and FooDB non-scientific. The FooDB non-scientific method uses a dictionary of common names of foods defined in the FooDB database, while the FooDB scientific method uses scientific names. Any entity that is extracted by these models is considered to be valid if
it is extracted by at least two of the three models that extract entities using common names (FoodNER, BuTTER or FooDB non-scientific), or
it is extracted by the FooDB scientific dictionary model, or
the FoodNER model has linked it to one of the external resources (FoodOn, SNOMED-CT or Hansard). In the case of the Hansard taxonomy, the linking can be accomplished by linking the entity to its parent category in the taxonomy (we refer to this scenario as the Hansard-Parent linking) or to the category that is most semantically similar to the entity (we refer to this scenario as the Hansard-Closest linking).
A postprocessing step was applied to remove food annotations that do not contain any nouns since these are more likely to be false positives and to remove words related to food that are too general to be useful or, more specifically, the words ‘food’, ‘foods’, ‘consumption’ and ‘drug’. The list is fixed and consists of these four words listed there. They were included since these words appear most often in the retrieved abstracts from PubMed and are related to the general entities (papers related to food consumption and also papers that have studied food–drug interactions).
We would like to emphasize that by applying the proposed annotation pipeline, we are actually generalizing the knowledge that is learned by BuTTeR and FoodNER, supplemented with the information stored in the other semantic resources, on new data. Both NER models are trained on recipe description data, which is a completely different style of writing from the scientific one. This means that the results will consist of false positives and false negatives that further require to be checked and validated by domain experts. By performing this step, we obtain a ‘silver standard’ of annotated scientific abstracts with food entities, which needs to be further validated by domain experts (as presented in the next subsection).
Step 3: Domain expert’s validation of the automatic annotations
For the acquisition of ‘gold standards’ for the annotated corpus, human validation of the annotations is required. To enable domain expert validation, we have upgraded the FoodViz tool. The upgrades have followed the same design patterns that have already been used to develop FoodViz. We have added two new features: (i) implementing the functionalities to correct the annotations originating from the food voting scheme and (ii) integrating the scientific abstracts annotated with the food annotation voting scheme in order to be validated by domain experts. The selected abstracts that are part of the CafeteriaSA corpus are available at http://foodviz.env4health.finki.ukim.mk/#/cafeteria.
Once the abstracts were uploaded to FoodViz, the domain expert validation was performed in two stages (i) using FoodViz at an in-person workshop by a team of researchers and (ii) checking the consistency of the assigned semantic tags.
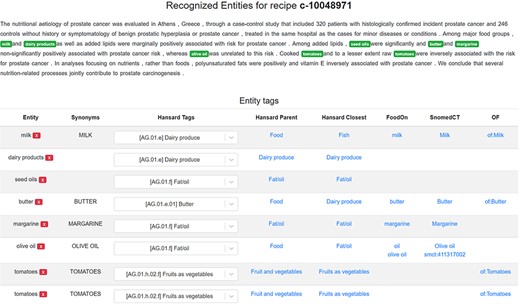
Figure 3 features an example of a scientific abstract annotated with food entities in the FoodViz tool. The experts are able to correct the highlighted food entities and their corresponding Hansard tags, while the matching to the FoodOn and SNOMED-CT ontologies is performed in an automatic manner. We need to point out here that the columns available in Figure 3 are presented from the previous goal of implementing the FoodViz tool. The FoodViz tool was used in this study to help the annotators to correct the false positives and true negatives. The Hansard tag is the tag that was produced by the food voting scheme, the Hansard Parent is the semantic tag that is in the higher level of the hierarchy and the Hansard Closest is the same as the Hansard tags or from some lower level from the hierarchy. They are produced by semantic similarity methods. In addition, the OF corresponds to OntoFood ontology that is used in the FoodOntoMap resource and can link the Hansard tags to tags from OntoFood. However, since OF contains a small coverage of food entities, it was not utilized in this study. In this study for the corrections, we are utilizing only the Hansard tag columns. The others are there only from the previous implementation of the FoodViz.
A scientific abstract annotated with food entities in the FoodViz tool.
Manually annotating a corpus is a task where the main challenge is to motivate a group of domain experts to be involved and trained to provide the annotations. In our case, a team of researchers (hereinafter referred to as annotators) was trained to correct the false annotations and to add annotations that were not recognized by our ML models. At first, a domain expert got familiar with the Hansard taxonomy and started with the annotation process to become acquainted with the selected abstracts and case-specific examples. After identifying the specific examples and possible dilemmas, domain experts prepared guidelines on how to (re)annotate the abstracts, which was important for the consistent use of semantic tags. The guidelines especially focused on errors’ avoidance and synchronizing the validation and re-annotation process. A team of 10 annotators met for an in-person interactive workshop, where they validated the automatically obtained annotations by removing the false positives and adding the false negatives. The workshop lasted ∼6 h, during which time each annotator curated 50 abstracts. The domain validation was performed using the semantic tags from the Hansard taxonomy.
In addition, it was important to differentiate between different types of foods that derive from the same food and annotate them with the appropriate tag. For example, ‘peanut oil’ was annotated as ‘fat/oil’, while ‘peanut butter’ was annotated as a ‘nut’ and not ‘fat/oil’ as it contains mainly peanuts. Overall, it was very important to understand the context of the abstract and tag or (re)annotate properly and not just blindly read and search for the food-related entities. If certain food could not be found in the Hansard corpus, the rule was to annotate it as a similar food or food group. For example, ‘pomelo’ cannot be found in the Hansard corpus, but since it is a citrus fruit, it should be annotated as such. In addition, if foods or food products could not be tagged because the tag does not exist in the Hansard corpus, they were tagged as ‘food’—a top-level category (e.g. Hansard corpus is lacking the category ‘seed’; therefore, pumpkin seeds, for example, were annotated as ‘food’). Moreover, the guidelines also emphasized that macronutrients should not be annotated (e.g. protein consumption), unless the macronutrient-related word is used as food or describes a type of food (e.g. coconut fat). Furthermore, processing methods were not annotated (e.g. cooked potato), with the exception of processing of edible oils (e.g. refined sunflower oil).
In the second stage of the validation process, two experts from the domain of food and nutrition were involved. They validated all the semantic tags in order to double-check the consistency of the assigned semantic tags. When necessary, errors were discussed among themselves and were corrected (tags removed or added) and entities were reannotated based on the rules in the guidelines.
Step 4: Alignment with other semantic resources—NCBO annotator
To make the annotated corpus available across different semantic resources, we utilized the NCBO annotator with two different semantic models/ontologies such as FoodOn and SNOMED-CT that are part of the BioPortal (36). For this purpose, the validated annotations were processed again two times with the NCBO annotator, each time with a different ontology. For each ontology, the annotation process was performed twice, once when we input the full abstract and once when we input just the food annotations one by one. In the end, we used the annotations that were obtained when each separate food annotation was passed through the NCBO annotator. The script used for the alignment is available at https://repo.ijs.si/matevzog/cafeteriancbo, where the configuration parameters used for the NCBO annotator are also presented.
Results and discussion
Following the annotation pipeline described earlier, first we used the food voting scheme to annotate the 500 selected abstracts. With this, we obtained the silver corpus that contains incorrectly extracted entities, i.e. false positives and false negatives. Next, the silver standard was imported into the FoodViz tool and used in an in-person workshop, where a team of researchers were trained for annotation guidelines by a domain expert and further corrected the false annotations. After that, two domain experts double-checked the annotated entities for consistency and corrected them if an inconsistency was found. As a final result, the corpus of 500 scientific abstracts has been refined with 6407 annotated entities with regard to Hansard taxonomy, 4299 for FoodOn and 3623 for SNOMED-CT, presenting the CAFETERIA gold standard. The difference that appears in the number of annotations per semantic resource is related to the coverage of the knowledge base of each semantic resource.
We need to point out here that the domain expert validation was performed only on Hansard semantic tags. Next, the food annotations available in the gold corpus were processed with the NCBO annotator to find their semantic tags with regard to SNOMED-CT and FoodOn. Table 1 presents the descriptive statistics for the number of annotated entities with regard to the three semantic resources used in our study. For this purpose, we provide the mean of annotated entities per abstract and the standard deviation of the number of annotated entities in the abstract. From the table, we can see that the Hansard corpus has the biggest coverage of entities, which indicates that the other semantic resources (SNOMED-CT and FoodOn) do not cover all food entities.
Descriptive statistics for the number of annotated entities from the three resources
| Semantic resource . | Mean . | Standard deviation . |
|---|---|---|
| Hansard | 12.78 | 6.67 |
| SNOMED-CT | 7.23 | 4.26 |
| FoodOn | 8.58 | 4.86 |
| Semantic resource . | Mean . | Standard deviation . |
|---|---|---|
| Hansard | 12.78 | 6.67 |
| SNOMED-CT | 7.23 | 4.26 |
| FoodOn | 8.58 | 4.86 |
Descriptive statistics for the number of annotated entities from the three resources
| Semantic resource . | Mean . | Standard deviation . |
|---|---|---|
| Hansard | 12.78 | 6.67 |
| SNOMED-CT | 7.23 | 4.26 |
| FoodOn | 8.58 | 4.86 |
| Semantic resource . | Mean . | Standard deviation . |
|---|---|---|
| Hansard | 12.78 | 6.67 |
| SNOMED-CT | 7.23 | 4.26 |
| FoodOn | 8.58 | 4.86 |
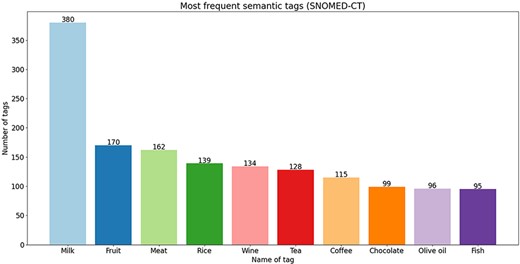
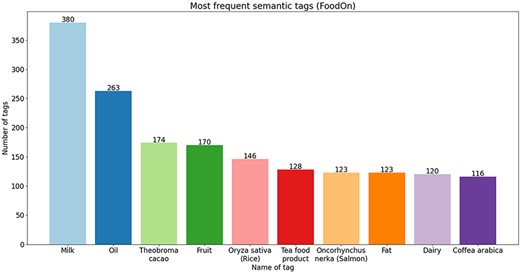
To go into more detail, in Figures 4–6, we present the 10 most frequent semantic tags from each semantic resource and the number of food concepts annotated with them. From the bar plots, we can see that there are tags that repeat themselves among the top 10 across the three semantic resources—with the ‘dairy produce’/‘milk’ tag being the tag with the most annotations from all three semantic resources. We should indicate here that these distributions are provided only for further utilization in new NLP tasks. For example, if we trained a new NER on this corpus, based on the distribution of the tags, we can know where the false discoveries can happen and for which tags good performance will be achieved since the distribution is not balanced. The distribution provides an overview of the coverage of the annotated corpus.
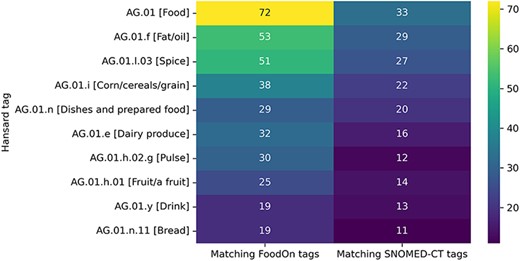
Figure 7 depicts the 10 semantic tags from the Hansard taxonomy that correspond to the highest number of unique semantic tags from the FoodOn and SNOMED-CT ontologies. As we can see, the top-level semantic tag food contains the most diverse entities since the food entities for which there was no better match in the Hansard taxonomy were linked to this tag. The remaining tags in the figure correspond to broad food categories that contain diverse entities, such as fats, oils, spices, cereals, dishes and prepared food.
Top 10 semantic tags from the Hansard taxonomy that correspond to the highest number of unique tags from the FoodOn and SNOMED-CT ontologies.
On average, the semantic tags from the Hansard taxonomy are linked to 19.29 unique text phrases. On the other hand, the semantic tags from the FoodOn and SNOMED-CT ontologies are linked to 2.93 and 3.56 unique text phrases on average, meaning that the semantic tags from the Hansard taxonomy contain more diverse entities and are more coarse-grained.
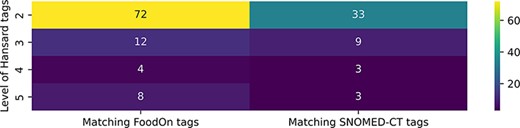
Figure 8 depicts the median number of unique semantic tags from the FoodOn and SNOMED-CT ontologies that correspond to each level of semantic tags in the Hansard taxonomy. It can be observed that the higher-level entities that describe more general concepts are mapped to a larger number of unique tags from the two ontologies, and this number generally decreases as we go into the deeper level of the taxonomy and the food entities become more specific.
Median number of unique FoodOn and SNOMED-CT semantic tags that are matched to the Hansard tags of each level of the taxonomy.
Figures 9 and 10 feature the food entities that are linked to some of the more diverse semantic tags from the Hansard taxonomy, more specifically the tags referring to fat/oil and spice, respectively. The size of each food phrase is proportional to its frequency in the corpus. As we can see from Figure 9, the diversity of the food phrases linked to the same fat/oil semantic tag is owed to the fact that oils and fats can be produced from different sources (canola oil, olive oil, coconut oil and rice oil), can be processed differently (refined and unrefined) and can sometimes not only be listed individually but also listed one after the other, with the word oil at the end (soybean, corn, sunflower and sesame oil). On the other hand, in Figure 10, we can see that the spice tag is linked to not only a variety of common names of spices but also their scientific names (Crocus sativus, Capsicum annuum and Coriandrum sativum).
Text phrases linked to the AG.01.f [Fat/oil] Hansard semantic tag.
Food phrases linked to the AG.01.l.03 [Spice] Hansard semantic tag.
During the curation process, several types of mistakes produced by the initial automatic annotation using the FoodNER model could be observed. A common mistake is producing a false-positive food entity when a word that is the name of a food entity in some contexts can have a different meaning in a different context (for instance, extracting turkey when the text was referring to the name of the country Turkey or extracting liver when the text was referring to it as a human body part). Another mistake is the extraction of food entities when they occur as a partial match of another type of entity. For instance, the entity milk was sometimes extracted when the word milk occurs as part of breast milk, human milk, rat milk or milk fever, while the entities nut and peanut were extracted as part of nut allergy or peanut allergy. Finally, partial matches or wrong linking was produced when several types of entities are listed as a group, with overlapping words. For instance, in the case when the phrase soybean, corn, sunflower and sesame oil occurs in the text, the NER models might extract soybean, corn and sunflower as separate entities and sesame oil as a separate entity, instead of figuring out that the word oil is shared across the entities and the text actually refers to soybean oil, corn oil, sunflower oil and sesame oil.
Human annotators were especially required to resolve the aforementioned cases, and a second round of checking by domain experts was needed to make the annotations consistent across the different annotators.
In contrast to annotating food recipes, the annotation of scientific text introduces difficulties due to the complexity of the text, the use of scientific food names and the inclusion of different types of entities, which further cause food entities to be confused with other entity types. In the text of food recipes, it is far less likely to encounter false positives of the aforementioned types. If the entities liver, turkey, milk, nut or peanut are mentioned in a food recipe, they usually refer to food entities since the entities they are commonly confused with (i.e. breast milk, human milk, rat milk, milk fever, peanut allergy, nut allergy or liver as a human organ and turkey as a country) are not commonly found in recipe text. The annotation of the scientific names of food entities, which are also not used in recipe text, requires additional efforts on the annotators’ part, especially when the annotators are not domain experts.
The CafeteriaSA corpus is available in the BioC format (37). The BioC format is well known and developed for sharing interoperable biomedical data that are further utilized for text mining experiments. We developed three different variants depending on the semantic resources used for annotating the food entities. The semantic resources are Hansard taxonomy (25, 26), SNOMED-CT (28) and FoodOn (27). The corpus can be accessed at https://zenodo.org/record/6683798#.YrLosexBwhl.
To show the utility of the existence of the annotated corpus, we further used it for generating an NER model intended specifically for the extraction of food entities from scientific text. We used a BERT model that was already pre-trained on large amounts of textual data and fine-tuned it for the NER task with the CafeteriaSA corpus. In this case, the NER task is treated as a classification problem where the classes are the tags from the IOB tagging scheme, i.e. the goal is to determine whether each word in the text is inside, outside or at the beginning of a food entity. We evaluate the model using cross-fold validation and report its performance in terms of the macro-averaged F1 score of all classes, averaged across all folds. Further details of the model’s training and evaluation procedures are beyond the scope of this paper; however, the methodology is similar to that of the FoodNER model.
We need to point out here that the food voting scheme with the models trained on the recipe dataset provided a macro F1 score of 0.58 evaluated on scientific text. On the other hand, the NER model obtained by fine-tuning BERT models on the CafeteriaSA corpus achieves macro-averaged F1 scores of 0.89, meaning that the corpus enabled an improvement of 0.31 in the extraction of food entities from scientific text. This result indicates that the CafeteriaSA corpus is beneficial for training the state-of-the-art NLP models for extracting food entities from textual data. In addition, it points out that the resources that will involve more different writing styles should also be researched and made available in future. Currently, the CAFETERIA project covers two different styles: recipe descriptions and scientific abstracts.
Conclusions
As a part of an EFSA-funded project, we have developed the first corpus of scientific abstracts annotated with food entities. For this purpose, an annotation pipeline supporting the following four steps has been introduced, in order to support (i) collecting a set of scientific abstracts; (ii) automated annotation, where food information extraction methods developed using recipe text data were used for automatic extraction of food entities from scientific text; (iii) domain validation of the automatically extracted entities; and (iv) making the corpus aligned with different food semantic resources. The annotations are available across the following semantic resources from the food and health domains: Hansard taxonomy, FoodOn and SNOMED-CT. The gold-standard corpus consists of 500 scientific abstracts, with a total of 6407 annotated entities with regard to Hansard taxonomy, 4299 for FoodOn and 3623 for SNOMED-CT. The coverage per semantic resource depends on the semantic tags that are available by each resource.
The existence of resources such as CafeteriaSA can facilitate several further research directions on the topic of food information extraction from textual data. First, they allow the development of NLP methods that can be used for the extraction of food information from textual data. With this, the new knowledge that is published in food science can easily be traced. Next, the extracted food entities can be linked to chemical, microbiome and other biomedical entities, which will allow training models for predicting and exploring the relations between them. Last but not least, CafeteriaSA has enriched the existing food-related semantic resources, so that they can be further used for investigating the links between food systems, human health and the environment.
The CafeteriaSA corpus is the first resource and as such it is still limited in the number of annotations. It covers a wide range of food entities; however, both the coverage of the semantic tags and the number of annotated entities should be enlarged. In the future, we aim to sample and collect abstracts for the low-representative semantic tags, preprocess them with the CAFETERIA pipeline and include them in the corpus in order to have more balanced distributions of the semantic tags. A potential direction for future work is also extending the corpus with annotations of the food entities with semantic tags from other resources; however, this requires human annotation experts who are familiar with the resources in question.
Conflict of interest
None declared.



![Text phrases linked to the AG.01.f [Fat/oil] Hansard semantic tag.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/database/2022/10.1093_database_baac107/3/m_baac107f9.jpeg?Expires=1716343840&Signature=2IsMh8pb70H9epzALYBoW~IbSxEMHAmzhN8jfAY7BxBRJd6uHW8MZ3kT8cqnjBFrSp5K68H6fc5N0rDo0WQur8rv0tzkB5k0Ngq5iI-8vv1ZEJpjjcQarTrBFt~81IRiJ5ZBWvnb0LGqyyzDWdYWr6BGFdLNGczjtOTOyYAuWaqZsxLZqaEHJx3EuA0J5xnNC2JmT0Coq69M6Wut0QZeHz6RrEwf-1Crx8ITAJvLtNi7aad5TY7f5MQ4BxayA1b33WqE6V6xKt3pdWOrPu8DNK6pwGDrAWnLF4Pi9CPk6anGjjRT53ULs0X5zHakhBMWnpx6~NALiUqOdtsjpag6ZA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Food phrases linked to the AG.01.l.03 [Spice] Hansard semantic tag.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/database/2022/10.1093_database_baac107/3/m_baac107f10.jpeg?Expires=1716343840&Signature=fqOPO9w9WjEADyiyF-p0gW5MqcXh-Hn4TWrGvlP6sbBaApZJfavz3hsmEfjS6MS2eRBBTMaJig3IapdT~IRJST2hc3oSfJkguugq~hu9NeUIlpIVzxnc9Xr0lsANcY41Uu5TQOc4dLFuJu1GKk0T5uiDbmaMEm7TwsxBL0oWbES9bo2gtZVV0CdN7voe9HskK7UrxObqSpcVLWTlKUo4jgJPk8YQUaG~NSuBzbmMF-NoWU0Ze575MM06CuwPxFti8xovUfGjke2tHDrrMYmm0uSCW0LzOI3N-YzbyQFBNeczqea7UESzsVezLQVhxcjH~QCHTMn0QkumnIUxPLOjQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
