-
PDF
- Split View
-
Views
-
Cite
Cite
Stanley J. F. Laulederkind, Mary Shimoyama, G. Thomas Hayman, Timothy F. Lowry, Rajni Nigam, Victoria Petri, Jennifer R. Smith, Shur-Jen Wang, Jeff de Pons, George Kowalski, Weisong Liu, Wes Rood, Diane H. Munzenmaier, Melinda R. Dwinell, Simon N. Twigger, Howard J. Jacob, RGD Team, The Rat Genome Database curation tool suite: a set of optimized software tools enabling efficient acquisition, organization, and presentation of biological data, Database, Volume 2011, 2011, bar002, https://doi.org/10.1093/database/bar002
Close - Share Icon Share
Abstract
The Rat Genome Database (RGD) is the premier repository of rat genomic and genetic data and currently houses over 40 000 rat gene records as well as human and mouse orthologs, 1771 rat and 1911 human quantitative trait loci (QTLs) and 2209 rat strains. Biological information curated for these data objects includes disease associations, phenotypes, pathways, molecular functions, biological processes and cellular components. A suite of tools has been developed to aid curators in acquiring and validating data objects, assigning nomenclature, attaching biological information to objects and making connections among data types. The software used to assign nomenclature, to create and edit objects and to make annotations to the data objects has been specifically designed to make the curation process as fast and efficient as possible. The user interfaces have been adapted to the work routines of the curators, creating a suite of tools that is intuitive and powerful.
Database URL:http://rgd.mcw.edu
Introduction
The pace and volume of genomic and genetic research has increased dramatically over the past decade due to technical advances in DNA sequencing and decreasing costs of such research. Many biological databases exist to organize and store either the sequencing data or associated biological data or both. Although the Rat Genome Database (RGD) does not store nucleotide or protein sequences, access to gene- and protein-specific sequence data is provided on each RGD gene report page by links to external databases such as Entrez Nucleotide and UniProtKB. Those links are imported via automated pipelines and are displayed as hyperlinked accession numbers on all RGD gene report pages.
Biocuration at RGD involves identifying data objects (QTLs and strains) in the literature or by direct interaction with researchers, assigning nomenclature to data objects (genes, QTLs, and strains), and annotating biological information to those data objects. Biological annotations are based on experimental data published in peer-reviewed journals. Currently, this work is done at RGD by seven full-time curators, five of whom have PhDs and two of whom have master’s degrees (1). Over the past few years RGD has made a concerted effort to improve the quality and quantity of manual biological curation through the use of ontologies, targeting of important data sets and improved curation software. An RGD team of biocurators and software developers has designed a web-based software suite that can download external data through pipelines, handle manual data curation and perform editing of any existing data. The components of the software suite include (i) a gene nomenclature tool, (ii) an object creation and editing tool and (iii) an ontology annotation creation and editing tool.
The curation tool suite
The gene nomenclature tool
Gene records in RGD are initially derived from NCBI’s gene records. An automated pipeline downloads gene data from Entrez Gene, compares the data to existing records in RGD and updates existing records or creates new records for genes which do not already exist in RGD. At that point it is the responsibility of RGD to assign official nomenclature to rat genes or confirm that official nomenclature has already been assigned to rat genes. As part of its role in assigning rat gene nomenclature, RGD works with Mouse Genome Informatics (MGI) (2) and the HUGO Gene Nomenclature Committee (HGNC) (3) to coordinate assignment of gene symbols and gene names for rat, mouse and human. HGNC is responsible for assigning official nomenclature to human genes and, in general, the mouse and rat nomenclature is assigned to match the nomenclature of human orthologs. The three groups communicate on an ongoing basis to ensure accuracy and consistency of nomenclature assignment. An ortholog file is generated at MGI, based on HomoloGene ortholog groups (4) and some manual editing of ortholog groups. This file is downloaded on a weekly basis and the ortholog relationships between rat, mouse and human genes are loaded into the RGD database by an automated pipeline. However, because the rat genome assembly is neither as complete, nor as well annotated as the human and mouse assemblies, RGD curators must also do some ortholog group editing to fill in gaps where the rat gene is missing from the MGI ortholog file. That ortholog group editing usually entails searching HomoloGene, BLASTing mRNA or protein RefSeq sequences, and checking synteny of the orthologs on genome viewers to determine which rat gene is the true ortholog of a particular ortholog group. Because of the extensive checking that is done prior to making these manual assignments, such changes are given priority in the RGD database.
The RGD Gene Nomenclature Tool utilizes the ortholog group data downloaded from MGI and edited at RGD to automatically generate lists of rat genes whose symbols or names differ from that of either mouse or human in the assigned ortholog group. This process is run each time a curator accesses the nomenclature tool. The user interface was built to be intuitive and fast enough to handle hundreds of nomenclature changes in a relatively short period of time.
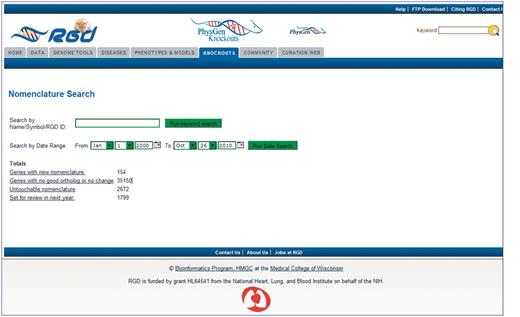
The homepage of the Gene Nomenclature Tool (Figure 1) displays results of the current ortholog pipeline download by category. The categories of ‘Genes with new nomenclature’ and ‘Genes with no good ortholog or no change’ are updated on a daily basis from the ortholog pipeline. ‘Genes with new nomenclature’ refers to genes with any change detected in the corresponding mouse or human ortholog. ‘Genes with no good ortholog’ refers to genes with no assigned ortholog from mouse or human and to genes where both mouse and human orthologs have nomenclature which is not appropriate for the rat gene, such as a ‘LOC####’-type temporary gene symbol. The status of the other two categories displayed (‘Untouchable nomenclature’ and ‘Set for review in next year’) depend on curator interaction with the pipeline specifications or with the editing portion of the tool. ‘Untouchable nomenclature’ is a category of genes for which it has been decided by nomenclature committee consensus that rat, mouse and human nomenclature will not be identical, usually because of problems of determining true orthologs between the species. This information is built into the pipeline that feeds the Gene Nomenclature Tool. ‘Set for review in next year’ consists of genes for which curators have decided to delay any nomenclature changes until sometime in the future. The delay is usually based on unresolved nomenclature discrepancies among the three species. A number count for each category is displayed to let the curators know the status of that category. The category names are hyperlinked to display pages where the curator can see all the genes or ortholog groups of that particular category. The search box gives the option of looking for a particular gene or family of genes.
Homepage of the Gene Nomenclature Tool. Pre-loaded categories are displayed as hyperlinks. Individual genes or families of genes can be searched by the keyword search box. Genes that are scheduled to be reviewed in the future can be searched through the date search function.
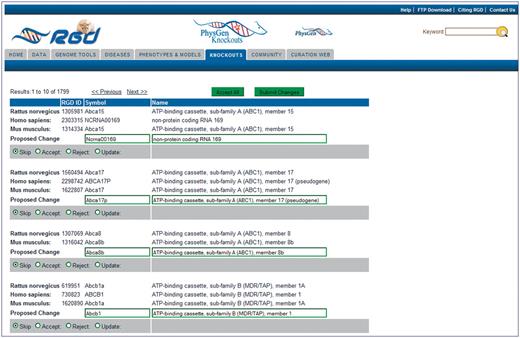
The editing display (Figure 2) shows the powerful nature of this tool. Ortholog groups are listed in alphabetical order, so changes to gene family members can be viewed at the same time. The tool automatically proposes an updated symbol and name for each gene based on the human ortholog nomenclature. If there is no human ortholog the proposed update is based on the mouse ortholog. The proposed changes appear in text boxes underneath each ortholog group. The text boxes are editable, which gives the curator an option to manually make a change to the nomenclature. For every proposed change the curator is given four radio button options: Skip, Accept, Reject or Update (change the next date of review). The layout of the ortholog groups on the page and the editing options allow one to rapidly review and update nomenclature in this tool. All nomenclature changes are tracked for each gene on the RGD report page of that particular gene. All previous symbols/names are retained as synonyms.
Editing page of the Gene Nomenclature Tool. All ortholog groups are listed in matched sets to simplify comparison of the rat nomenclature to the mouse and human nomenclature. The proposed change/editing text boxes and action selection radio buttons are at the bottom of every ortholog group. An ‘Accept All’ button is located at the top and bottom of every editing page so that all proposed changes on the page can be approved at once if the curator decides they are acceptable. ‘Submit Changes’ buttons are located next to the ‘Accept All’ buttons to send the changes to the database.
The object creation and editing tool
The Object Creation and Editing Tool allows the curators to add new data objects (QTLs or strains) to the database, and edit database records for existing data objects (QTLs, strains or genes). The first part of adding data in the Object Creation and Editing Tool for new QTLs and strains is assigning nomenclature. New QTLs or strains are determined by curator search of the biomedical literature in PubMed or by direct data submission to RGD from a researcher. As in the case for gene nomenclature, the names assigned to QTLs are made according to the ‘Guidelines for Nomenclature of Genes, Genetic Markers, Alleles and Mutations in Mouse and Rat’ (5), while the names assigned to strains follow the ‘Rules for Nomenclature of Mouse and Rat Strains’ (for a quick guide to all nomenclature rules, Ref. 6). Since QTLs that are assigned the same trait are given the same root symbol followed by a sequential number, the tool has a symbol search function which shows the curator all symbols with the same root. That allows the curator to easily assign the next sequential number for that symbol root. The symbol search works similarly for strain nomenclature, as substrains have symbols and names that include parts of the parent symbol and name. The symbol search lists all related strain symbols, which aids in assigning a symbol to a new strain record. Another important nomenclature feature of the Object Creation and Editing Tool is the ability to add synonyms and unofficial symbols to the object records. This allows the object records to be searched with all the various names that may be used in the literature.
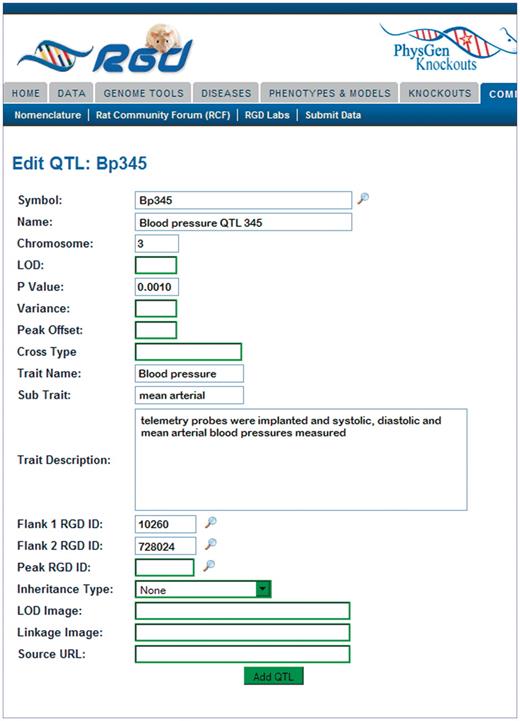
The tool provides separate data entry templates for QTLs and strains to accommodate the differences in data relevant to the particular object type. For QTLs the tool template allows, in addition to nomenclature, entry and editing of genomic data (chromosome number, upstream and downstream flanking markers, peak marker), trait data (trait and subtrait names from a controlled vocabulary being developed at RGD, trait description), statistics (LOD score, P-value, variance) and source information (Figure 3). For strains the tool template allows, in addition to nomenclature, entry and editing of genetic/genomic data (genetic markers, chromosome alterations), breeding data (type, inbred generation number, origin) and source information. For gene records the tool provides editing of nomenclature, object status and curation notes.
The Object Creation and Editing Tool: QTL entry page. The QTL entry page consists of many text boxes specific for the different types of information collected by the curator to describe a QTL. Clicking the search icon to the side of the symbol box generates a pop-up window where current QTL symbols may be searched.
While dealing with multiple QTLs or strains with similar characteristics, the tool can be used to ‘clone an object’. This allows creation of an additional object with attached data, which needs only minimal changes (like name, statistical values and source) to complete the new object record. Through a similar process data can be imported into the tool for any QTL, strain or gene in RGD to allow for editing of errors or any changes that may be necessary for any particular data object. At times, there is also a need to simultaneously change the status of multiple objects, due to user request or a new assembly of the rat genomic sequence. The ‘Object Status’ function of the tool was designed to handle these types of multiple updates.
The ontology annotation creation and editing tool
All the manual biological curation of data objects at RGD begins with object-, species- and topic-specific PubMed searches by the curators. Since RGD curates biological properties of genomic objects (genes, QTLs) and other data objects (rat strains), the Ontology Annotation Creation and Editing Tool was designed to accommodate all of those elements. The tool utilizes five different ontologies or vocabularies: Gene Ontology (GO), Disease Ontology (the C branch of MeSH), Pathway Ontology (PW, an ontology developed at RGD), Mammalian Phenotype Ontology (MP from Mouse Genome Informatics), and Behavior Ontology (the F branch of MeSH). The tool combines data objects, ontology terms, references and other supplementary data to allow curators to make various types of annotations.
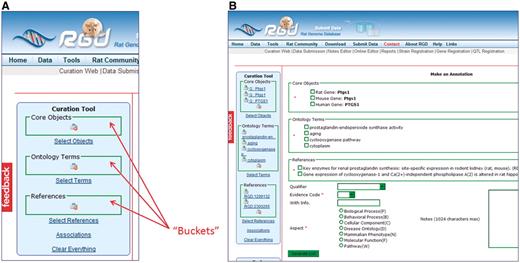
The key feature of the RGD Ontology Annotation Creation and Editing Tool is the use of ‘buckets’ to hold selected core objects (genes, QTLs and strains), ontology terms and references (Figure 4A). Each bucket has its own search function, specific for the category of items which that bucket holds. Each bucket can hold multiple items, which allows for construction of annotations using a variety of item combinations and enables curators to make multiple annotations simultaneously.
(A) The Ontology Annotation Creation and Editing Tool homepage has ‘buckets’ for holding selected items. Each bucket (core objects, ontology terms and references) has its own search function, which appears in the right frame of the page when ‘Select Objects’, ‘Select Terms’ or ‘Select References’ is clicked. (B) Annotation frame. All the items from the buckets are repeated in the annotation frame to allow them to be selected for annotations. More information choices are available below the bucket items as drop-down text boxes for qualifier terms and evidence codes, radio buttons for ontology aspect and text boxes for free text information.
The ‘core object’ search allows searching for individual object types (genes, QTLs, strains), or combinations of object types, with functionality that includes autocomplete and drop-down options of ‘Equals’, ‘Contains’, ‘Begins with’ or ‘Ends with’. The ontology term search also features autocomplete, as well as the option to choose any or all of the available ontologies (Gene Ontology, Disease Ontology, Pathway Ontology, etc.). Both object and term searches return results in alphabetical order with exact matches listed first. Each individual result can be transferred to the appropriate bucket by the click of a selection icon.
The reference search allows searching by RGD ID, PubMed ID, author, year or keyword. If the search is for something other than an ID, the results can be ordered by year, citation or title. Most of the time the reference search involves PubMed ID entry. If the PubMed ID entered in the tool is not yet stored in RGD, the tool will automatically download the associated abstract from the PubMed website (7), assign an RGD ID to the abstract, and put the resulting RGD ID into the reference bucket.
After at least one item occupies each of the buckets, an option can be chosen to make an annotation(s) with the selected items. All of the items from the buckets get transferred to the ‘annotation’ frame of the tool, where any of those items may be selected for use in constructing an annotation. The mixing and matching of different items from the buckets allows for a great variety of possible annotations. Additional text fields, including qualifier and evidence code drop-down menus, and radio buttons for ontology aspect allow the curator to add more information to the basic data defined by the items in the buckets (Figure 4B).
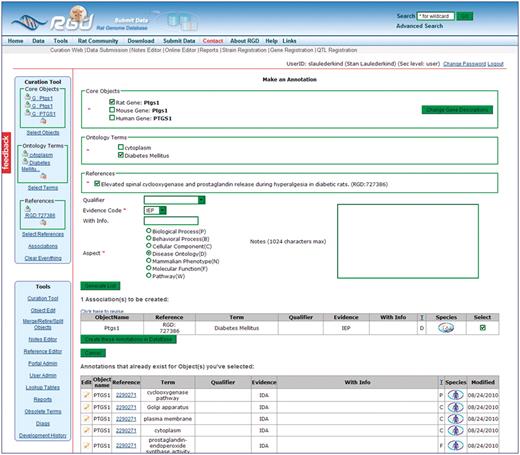
Clicking the ‘Generate List’ button results in an annotation(s) being displayed in an intermediate screen (Figure 5) where editing may be done before committing the newly generated annotation(s) to the database. Errors can be corrected or changes can be made as easily as the original annotation(s). The same annotation frame holds all pre-existing database annotations for items in the core object bucket. This gives a visual aid to curators, so they know what has already been curated for the selected core objects. Each annotation is also connected to an annotation editing tool, where they can be altered after they have been committed to the database. This gives curators the ability to edit every component of an annotation both before and after committing that annotation to the database.
This intermediate annotation page holds the currently constructed annotation for Ptgs1, all existing annotations for objects in the Core Objects bucket (PTGS1/Ptgs1 for human, mouse and rat), and options for editing. The current annotation can be edited by reselecting items above the annotation and re-clicking the ‘Generate List’ button. Any pre-existing annotation at the bottom of the frame can be edited by clicking the edit icon to the left of the annotation.
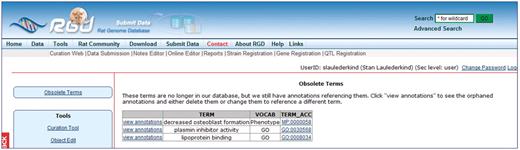
Another editing-related feature of the curation tool is the monitoring of obsolete terms in the Gene Ontology and the Mammalian Phenotype Ontology. During the weekly pipeline load of ontologies to RGD, the curation tool checks to see if any terms have been declared obsolete in the individual ontologies. All obsolete terms, for which RGD has annotations, are listed in the curation tool (Figure 6) with links to the annotations containing those terms, so a curator can edit or delete those annotations. The list has a real-time update, so as soon as annotations with obsolete terms are corrected in the database, the list is remade to reflect the corrections.
A list of obsolete ontology terms with links to annotations containing those terms.
Software development
The gene nomenclature tool
The Gene Nomenclature Tool is built on J2EE (8) technologies and driven off the RGD Oracle database. The tool is built to run on any Java container that implements the Servlet 2.4 and JSP 2.0 specification or above. The Spring (9) framework is used for dependency injection at run time and is configured to house regular expression patterns that identify nomenclature that will or will not be processed in the user interface. A pagination algorithm has been added to display results in groups of 10 in addition to a directory of all pages in the result set. This allows curators to move through large result sets efficiently. The user interface relies heavily on recent improvements in DOM technology along with CSS (cascading style sheets). Supported browsers include IE 8+, Firefox 3+ and Safari 5+.
The Object Creation and Editing Tool
The Object Creation and Editing Tool is built on J2EE technologies and driven off the RGD Oracle database. It is a CRUD (create, read, update, delete) application built to run on any Java container that implements the Servlet 2.4 and JSP 2.0 specification or above. The web application is built on the Spring framework’s MVC (model-view-controller) architecture. The tool utilizes AJAX (10) allowing the curator to update fields without a refresh of the page. In addition, an AJAX quick lookup has been included to allow for quick translation from symbol to RGD ID without the need for a new page. New objects and updates to existing objects are run through a validation layer to reduce the probability that errors make it into the database. Supported browsers include IE 8+, Firefox 3+ and Safari 5+.
The ontology annotation creation and editing tool
The Ontology Annotation Creation and Editing Tool was developed in PHP (11) and is driven off the RGD Oracle database. It is built on a MVC architecture that includes view helpers for AJAX, JavaScript and HTML forms. In addition, built-in validation routines are included to assist curators in quality control. The curation software templating system allows for new software features to be integrated seamlessly. The user interface is built on standard web technologies including HTML, JavaScript and CSS. The ‘obsolete terms’ feature of the tool is based on automatic downloads of OBO (Open Biological and Biomedical Ontologies) files (12,13) via FTP and HTTP. After parsing of the terms to determine differences between new and old ontology files, an SQL query determines which annotations use obsolete terms.
Recent updates to the Ontology Annotation Creation and Editing Tool involve various alterations on the back-end to improve functionality of the user interface. To implement fully automated PubMed ID importing, the existing CGI program that handles abstract download and RGD ID assignment has been modified so that the CGI program can accept the PubMed ID from the curation tool automatically and send the result back to the curation tool. To put the ontology term search results in alphabetical order with exact match listed first, a scoring function has been applied to the SQL query. To make better use of HTML anchors so the browser can automatically scroll to a desired position in a web page in the tool, the PHP framework has been modified. As with the other tools, supported browsers include IE 8+, Firefox 3+ and Safari 5+.
The architecture and capabilities of the RGD curation software may be of interest to the informatics community. However, because the curation tool software was designed specifically to fit the RGD database schema, it would not be easily adaptable for use with other databases and so it has not been made publicly available.
Summary
The Rat Genome Database curates many types of data (disease, phenotype, pathway, molecular function, biological process, cellular component and nomenclature) for a variety of objects (genes, QTLs, strains). To perform that biocuration efficiently, the development of a suite of software tools was necessary. To best match the software to the curation process, the tools were designed according to the curators’ specifications. Development of the Gene Nomenclature Tool has provided a powerful and efficient way to keep abreast of changes in the nomenclature of orthologous genes in mouse and human. The Object Creation and Editing Tool provides a variety of functions essential to the curation of QTLs and strains. It also has the versatility of being able to edit basic information for genes. The Ontology Annotation Creation and Editing Tool is another multifunctional tool whose main use is assigning biological annotation to data objects. It has been fine-tuned after multiple rounds of use, re-evaluation, and design upgrades. Performing together, these software tools provide a robust and efficient curation process that RGD has used to deal with an enormous and constantly growing amount of genetic/genomic and biological information.
Funding
National Heart, Lung and Blood Institute on behalf of the National Institutes of Health [HL64541]. Funding for open access charge: National Heart, Lung and Blood Institute on behalf of the National Institutes of Health [HL64541].
Conflict of interest. None Declared.